Inflammatory Gene Expression And Nuclear Factor
Most inflammatory genes are regulated by the actions of proinflammatory transcription factors such as NF-κB,81,82 although the action of NF-κB is modulated by the status of other transcription factors and signaling pathways including STATs, AP-1, and the p38 MAPK pathway. These control NF-κB activity either by protein-protein interaction or by posttranslational modifications of NF-κB subunits. NF-κB is also involved in the regulation of the innate and adaptive immune system, cell cycling, and proliferation and repair processes.81,82
Members of the Rel family and of the NF-κB family act as homodimers or heterodimers to form NF-κB. These native proteins are generally cleaved to shorter active DNA-binding proteins .81,82 In contrast, Rel proteins possess a C-terminal transcription activation domain that mediates transcription. Therefore each member of the NF-κB family must associate with an Rel family member to enable gene transcription to occur.81,82
NF-κB activity is tightly controlled to prevent unconstrained inflammation and maintain homeostasis.81,82 This process requires several layers of negative feedback loops such as NF-κB-induction of its inhibitory proteins IκBα and A20. IκBα shuttles NF-κB out of the nucleus, whereas A20 deubiquitinates several components of the NF-κB activation pathway, ultimately resulting in a loss of IκBα degradation.81,82
Mohammad Yaseen Sofi, … Khalid Z. Masoodi, in, 2022
The Processes Involved Are:
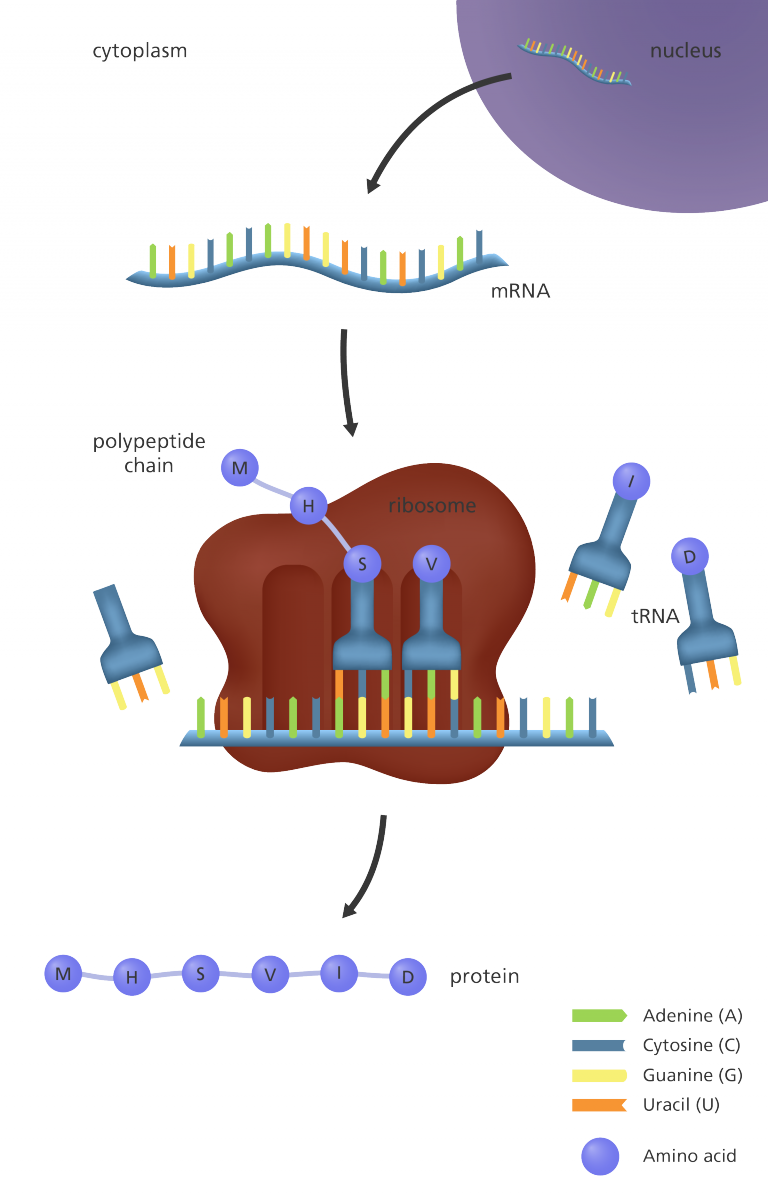
- Most of the mature mRNAs produced after modifications are transported from the nucleus to the cytoplasm where the next step in gene expression takes place.
- This is achieved by moving the mRNAs through tiny pores in the nucleus to reach the cytosol.
- The sequence in the mRNA is translated into a protein with the help of several components such as ribosomes, tRNAs or transfer RNAs, and enzymes called aminoacyl tRNA synthetases.
- Translation of mRNA involves 3 important steps initiation, elongation, and termination, leading to the formation of polypeptide chains.
- In this final step, the polypeptide chains or random coils formed during translation fold into a 3D structure giving rise to a functional protein.
- Failure to fold leads to protein inactivity and misfolded proteins have abnormal functionalities compared to correctly folded ones.
- Also, proteins can be modified by various methods such as phosphorylation, glycosylation, ADP ribosylation, hydroxylation, and addition of other groups.
Ivgene Transfer In The Cns
Reporter genes, such as -galactosidase and luciferase, have played vital roles in the understanding of the molecular mechanisms of gene expression. The concept of a reporter gene provides a much simpler method for genetic analysis than the conventional hybridization techniques. A defined nucleotide sequence is introduced into a cell, whose expression results in a well-defined protein, which can be measured using the reporter probe. These reporter genes also have provided important information in the design of delivery systems in gene transfer into somatic tissue. However, monitoring these reporter systems in living animals requires either highly invasive tissue biopsy or post-mortem analysis of the animal under study. Optical imaging techniques, using GFP and luciferase, have been developed to provide non-invasive monitoring of gene expression in vivo, although these techniques are limited to either organisms which are transparent to light, or, in larger animals, to detection of gene expression near the surface of the skin . In addition, accurate quantification of the emitted light is difficult due to absorption of the light by the animal. Detection in the CNS is further compromised by the inability of light to penetrate the skull. Therefore, techniques are required which enable repetitive, non-invasive, quantitative imaging of reporter genes in living animals, which should be equally applicable to human studies.
H.D. Jones, in, 2003
Recommended Reading: Kuta Software Similar Triangles Answer Key
Dna Methylation And Demethylation In Transcriptional Regulation
DNA methylation is a widespread mechanism for epigenetic influence on gene expression and is seen in bacteria and eukaryotes and has roles in heritable transcription silencing and transcription regulation. Methylation most often occurs on a cytosine . Methylation of cytosine primarily occurs in dinucleotide sequences where a cytosine is followed by a guanine, a CpG site. The number of CpG sites in the human genome is about 28 million. Depending on the type of cell, about 70% of the CpG sites have a methylated cytosine.
Methylation of cytosine in DNA has a major role in regulating gene expression. Methylation of CpGs in a promoter region of a gene usually represses gene transcription while methylation of CpGs in the body of a gene increases expression.TET enzymes play a central role in demethylation of methylated cytosines. Demethylation of CpGs in a gene promoter by TET enzyme activity increases transcription of the gene.
Toxicogenomics For Compound Classification And Prioritization
Gene expression signatures have also been used to classify compounds into different toxicity classes based on mechanism and target organ specificity or potency. Statistical techniques, such as discriminant analysis, Bayesian classification, and neural networks, are used to build prediction models that automatically classify compounds in different classes based on gene expression signature correlations. Such classifications can be useful to prioritize compounds for further testing , which improves the efficiency of the assessment paradigm. The gene expression responses in biological systems have been collected in public databases such as the Comparative Toxicogenomics Database , Environment, Drugs, and Gene Expression database , and Chemical Effects in Biological System knowledge base. These databases provide a useful tool for the analysis and comparison of microarray data and facilitate the identification of gene expression signatures related to environmental exposures.
-
Correlation mode: a correlation map based on average gene expression profiles and
- 3.
-
Gene Finder mode: a differential search tool that allows users to search for enhanced gene expression at the voxel level rather than at the structure level.
Figure 3.4. Anatomic Gene Expression Atlas .
Matthew T. Olson, Martha A. Zeiger, in, 2016
Read Also: Paris Jackson Father
Transcriptional Control Of Gene Expression
Transcriptional regulation is control of whether or not an mRNA is transcribed from a gene in a particular cell. Like prokaryotic cells, the transcription of genes in eukaryotes requires an RNA polymerase to bind to a promoter to initiate transcription. In eukaryotes, RNA polymerase requires other proteins, or transcription factors, to facilitate transcription initiation. Transcription factors are proteins that bind to the promoter sequence and other regulatory sequences to control the transcription of the target gene. RNA polymerase by itself cannot initiate transcription in eukaryotic cells. Transcription factors must bind to the promoter region first and recruit RNA polymerase to the site for transcription to begin.
The Promoter and Transcription Factors
In eukaryotic genes, the promoter region is immediately upstream of the coding sequence. This region can range from a few to hundreds of nucleotides long. The length of the promoter is gene-specific and can differ dramatically between genes. The longer the promoter, the more available space for proteins to bind. Consequently, the level of control of gene expression can differ quite dramatically between genes. The purpose of the promoter is to bind transcription factors that control the initiation of transcription .
Figure 17.10
Enhancers and Repressors
Control Of Gene Expression
- The structural genes contain the code for the proteins products that are to be produced. Regulation of protein production is largely achieved by modulating access of RNA polymerase to the structural gene being transcribed.
- The promoter gene doesn’t encode anything it is simply a DNA sequence that is initial binding site for RNA polymerase.
- The operator gene is also non-coding it is just a DNA sequence that is the binding site for the repressor.
- The regulator gene codes for synthesis of a repressor molecule that binds to the operator and blocks RNA polymerase from transcribing the structural genes.
The operator gene is the sequence of non-transcribable DNA that is the repressor binding site. There is also a regulator gene, which codes for the synthesis of a repressor molecule hat binds to the operator
- Example of Repressible Transcription: E. coli need the amino acid tryptophan, and the DNA in E. coli also has genes for synthesizing it. These genes generally transcribe continuously since the bacterium needs tryptophan. However, if tryptophan concentrations are high, transcription is repressed by binding to a repressor protein and activating it as illustrated below.
Recommended Reading: Abiotic Science Definition
What Is Gene Expression Analysis
Gene expression analysis is most simply described as the study of the way genes are transcribed to synthesize functional gene products functional RNA species or protein products. The study of gene regulation provides insights into normal cellular processes, such as differentiation, and abnormal or pathological processes.
Gene expression workflow.
Researchers may perform gene expression analysis at any one of several different levels at which gene expression is regulated: transcriptional, post-transcriptional, translational, and post-translationalprotein modification.
Transcription, the process of creating a complementary RNA copy of a DNA sequence, can be regulated in a variety of ways. Transcriptional regulation processes are the most commonly studied and manipulated in typical gene expression analysis experiments.
The binding of regulatory proteins to DNA binding sites is the most direct method by which transcription is naturally modulated. Alternatively, regulatory processes can also interact with the transcriptional machinery of a cell. More recently, the influence of epigenetic regulation, such as the effect of variable DNA methylation on gene expression, has been uncovered as a powerful tool for gene expression profiling. Varying degrees of methylation are known to affect chromatin folding and strongly affect accessibility of genes to active transcription.
What Is Gene Expression In Biology Quizlet
4.5/5gene expressiongeneticgenesin-depth answer
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as transfer RNA or small nuclear RNA genes, the product is a functional RNA.
Also, when a gene is expressed it is? Gene expression is the process by which the instructions in our DNA are converted into a functional product, such as a protein. It acts as both an on/off switch to control when proteins are made and also a volume control that increases or decreases the amount of proteins made.
In this manner, what is gene expression quizlet?
gene expression. the activation or “turning on” of a gene that results in transcription and the production of mRNA. genome. the complete genetic material contained in an individual. structural gene.
What is the final product of gene expression quizlet?
It is the process of converting the message from mRNA into a protein. Translation occurs in the cytoplasm at the ribosomes. The final product of translation, and gene expression, is a protein.
Don’t Miss: How Was Ancient Greek Civilization And Culture Affected By Geography
Mechanisms Of Gene Regulation Include:
- Regulating the rate of transcription. This is the most economical method of regulation.
- Regulating the processing of RNA molecules, including alternative splicing to produce more than one protein product from a single gene.
- Regulating the stability of mRNA molecules.
- Regulating the rate of translation.
- Transcription factors are proteins that play a role in regulating the transcription of genes by binding to specific regulatory nucleotide sequences.
Regulation Of Gene Expression By Small Rnas
mRNAs can also adopt structures that act as sensors for small regulatory RNAs that affect gene expression via RNARNA interactions . Anti-sense regulatory RNAs can be divided into cis-acting and trans-acting regulatory molecules. Cis-acting RNAs are antisense RNAs, transcribed from the opposite DNA strand and show perfect bp complementarity to the mRNA target. They can vary in size from a few tens to thousands of nucleotides and their abundance in different bacteria varies extensively . Anti-sense RNAs can affect transcription in essentially two manners: transcription interference or transcription attenuation . Transcription interference occurs when transcription initiated from one promoter is restrained by the action of a second promoter in cis. Attenuation is a mechanism that causes premature termination of transcription. Base pairing of antisense RNA to the cognate mRNA may induce the formation of a Rho-independent terminator, resulting in an aborted non-functional transcript, or in contrast, induce the formation of an anti-terminator structure thus allowing production of a full-length functional transcript. Another potential mode of regulation by antisense RNAs is to either promote or inhibit degradation of the target mRNA by endo- or exoribonucleases. In bacteria, the endoribonucleases RNase III and RNase E have for instance been linked to RyhB antisense RNA-induced target cleavage of sodB mRNA, encoding superoxide dismutase B .
Don’t Miss: Geometry Basics Segment Addition Postulate
Ii2 The Central Dogma
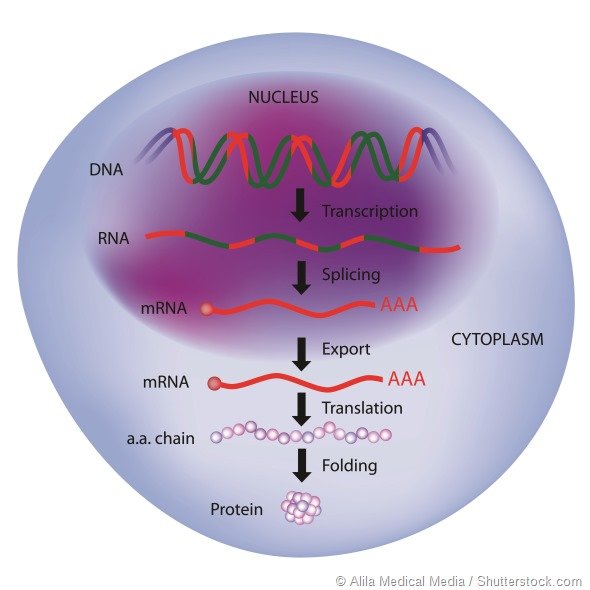
Genetic instructions or genes in the form of a sequence of DNA are first transcribed into mRNA and then translated into a protein. This process is often referred to as the central dogma.
Figure II.2 The central dogma of gene expression.
Recall that DNA consists of a long sequence of nucleotides. Each nucleotide contains a sugar , a phosphate group, and a nitrogenous base. There are four nitrogenous bases: adenine , thymine , guanine , and cytosine . The sequence of A, T, G, C ultimately determines the sequence of the RNA and protein that will be produced. A set of three nucleotides constitutes a codon. Each codon is translated into an amino acid .
If you wish to review these concepts , read this excellent review.
DNA is double stranded, resulting from complementary bases forming hydrogen bonds . So every strand of DNA contains a sequence of base pairs. You can see that DNA is a repository of information. This information is not stored in the binary language like in our computers, but with an alphabet of four bases .
Figure II.3 DNA structure.
Madeleine Price Ball, Wikimedia commons, CC0 1.0
- eukaryote
- The sequence of base pairs that encodes amino acids and is translated into a protein.
- splicing
- The process of cutting out introns and joining the exons of a precursor mRNA before it is sent for translation. See also: intron and exon.
Figure II.4 Transcription and translation.
Transcriptional Regulation In Learning And Memory
In a rat, contextual fear conditioning is a painful learning experience. Just one episode of CFC can result in a life-long fearful memory. After an episode of CFC, cytosine methylation is altered in the promoter regions of about 9.17% of all genes in the hippocampus neuron DNA of a rat. The hippocampus is where new memories are initially stored. After CFC about 500 genes have increased transcription and about 1,000 genes have decreased transcription . The pattern of induced and repressed genes within neurons appears to provide a molecular basis for forming the first transient memory of this training event in the hippocampus of the rat brain.
In particular, the brain-derived neurotrophic factor gene is known as a “learning gene.” After CFC there was upregulation of BDNF gene expression, related to decreased CpG methylation of certain internal promoters of the gene, and this was correlated with learning.
You May Like: Prince Jackson Real Father
Epigenetic Control Fo Gene Expression
The first level of control of gene expression is epigenetic regulation. Epigenetics is a relatively new, but growing, field of biology.
Epigenetic control involves changes to genes that do not alter the nucleotide sequence of the DNA and are not permanent. Instead, these changes alter the chromosomal structure so that genes can be turned on or off. This level of control occurs through heritable chemical modifications of the DNA and/or chromosomal proteins.
One example of chemical modifications of DNA is the addition of methyl groups to the DNA, in a process called methylation, In general, methylation suppresses transcription. Interestingly, methylation patterns can be passed on as cells divide. Thus, parents may be able to pass on the tendency of a gene to be expressed in their offspring. Other heritable chemical modifications of DNA may also occur.
Modification of Histone Proteins is an Example of Epigenetic Control
Figure 17.7
The first level of organization, or packing, is the winding of DNA strands around histone proteins. Histones package and order DNA into structural units called nucleosome complexes, which can control the access of proteins to the DNA regions . Under the electron microscope, this winding of DNA around histone proteins to form nucleosomes looks like small beads on a string . These beads can move along the string and change the structure of the molecule.
Figure 17.9
S In Protein Synthesis
Just like the process of transcription, the translation process also consists of three major steps initiation, elongation, and termination. A brief detail of these steps is as follows.
The process of translation begins with the assembly of the translational machinery. A specific codon, called initiation codon, is located at the beginning of mRNA. It signals the ribosomes to start the process of translation. The initiation codon codes for methionine amino acid.
A molecule of mRNA is recognized by a small ribosomal subunit via a specific nucleotide sequence located upstream the initiation codon. In prokaryotes, a specific purine-rich sequence is upstream the initiation codon called Shine-Dalgarno sequence. However, this sequence is absent in eukaryotes. The eukaryotic ribosomes recognize mRNA through its keep located several nucleotides upstream the initiation codon.
The small ribosomal subunit recognizes and binds to the mRNA and begins the screening process till it reaches the initiation codon.
The initiation codon is exposed at the P site of the ribosome. This AUG code is recognized by a special tRNA, called initiator tRNA, carrying the methionine amino acid. This is the only tRNA that directly goes to the P site of ribosomes.
Once the initiator tRNA has attached, the larger ribosomal subunit joins the complex resulting in the formation of a fully functional ribosome, ready to carry out protein synthesis.
You May Like: Algebra Nation Section 1 Test Answers