Residence Time Of Receptorligand Complexes And Its Effect On Biological Function
*Biochemistry
Publication History
Article Views are the COUNTER-compliant sum of full text article downloads since November 2008 across all institutions and individuals. These metrics are regularly updated to reflect usage leading up to the last few days.
Citations are the number of other articles citing this article, calculated by Crossref and updated daily. Find more information about Crossref citation counts.
The Altmetric Attention Score is a quantitative measure of the attention that a research article has received online. Clicking on the donut icon will load a page at altmetric.com with additional details about the score and the social media presence for the given article. Find more information on the Altmetric Attention Score and how the score is calculated.
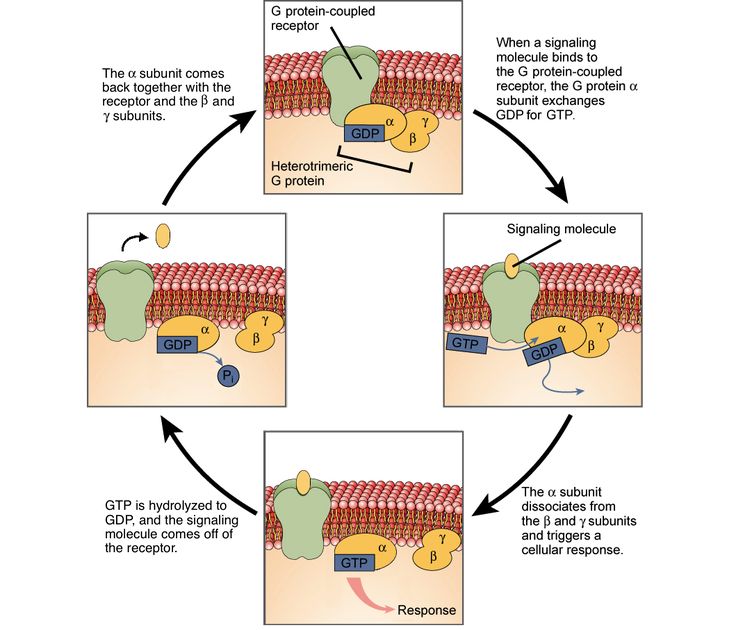
Practical Relationships Between Kd And
For the same concentration of added ligand and receptor, more complex formation will occur with a smaller value of Kd
Example 6.3.1
Receptor and ligand are added to a total concentration of 1.0 x 10-4M. The Kd for the RL complex is 1.0 x 10-4M. What is the concentration of , and at equilibrium?
The added ligand, L, will be partitioned between free and bound forms:
Ltotal = L + RL = 1.0 x 10-4M
L = 1.0 x 10-4M – RL
Similarly, the added receptor, R, will be partitioned between free and bound forms:
Rtotal = R + RL = 1.0 x 10-4M
R = 1.0 x 10-4M – RL
Substituting these values of L and R into the expression for Kd yields:
This is a quadratic with values a = 1, b = -3.0 x 10-4 and c = 1.0 x 10-8. This yields two possible values for RL:
RL = 2.62 X 10-4M or
RL = 3.82 X 10-5
Since the maximum possible value for is 1.0 x 10-4 , the first result is not possible and = 3.82 X 10-5M
Thus, 38.2% of the added receptor is in the complex formation x100%
Example 6.3.2
Same amount of added R and L, but this time the Kd for the RL complex is 1.0 x 10-6M. What is the concentration of , and at equilibrium?
his is a quadratic with values a = 1, b = -2.01 x 10-4 and c = 1.0 x 10-8. This yields two possible values for RL:
RL = 1.11 X 10-4M or
RL = 9.05 X 10-5
Since the maximum possible value for is 1.0 x 10-4 , the first result is not possible and = 9.05 X 10-5M
Thus, 90.5% of the added receptor is in the complex formation x100%
Interpretation Of Receptor Functionality
Ligand binding and second messenger screening assays can be utilized to ensure that mutant or chimeric GPCRs are functional. The relative expression levels determined by binding may vary however, ligand affinity should not be altered drastically. If ligand binding is drastically altered, consider using a different ligand . In a similar manner, MAP kinase activation may not occur or be relevant to a specific GPCR, thus another second messenger assay should be considered . Prior to going forward with trafficking analysis, all mutant or chimeric GPCRs should retain ligand binding and second messenger activation to demonstrate functionality .
Figure 9.2. Functional screening of 2C AR mutations by MAP kinase. An immunofluorescent assay for MAP kinase in response to agonist allows for screening of GPCR chimeras and mutations to ensure pharmacological function prior to further trafficking analysis. Permeabilized cells were stained with monoclonal antibody 16B12 to examine total cellular HA-2 AR expression. Note minimal pERK staining in the absence of agonist. Both 2C5-15 and 2CA7D ARs can activate MAP kinase and bind RX-821002 suggesting that they are not misfolded and are functional.
L.E. Goldfinger, in, 2013
Don’t Miss: What Do You Study In Psychology At University
Lentiviral Production And Transduction
For screening, 4×107 HEK293-V2M cells were transduced to achieve between 25 and 30% BFP-positive cells . An MOI of 0.3 ensured that the majority of infected cells receive one virus per cell. Transduced cells were sorted on day 2 post-transduction on a MoFlo XDP cell sorter and BFP-positive cells collected. Between 1.01.6×107 cells were collected for each transduction and maintained in media supplemented with 2g/mL puromycin to maintain lentiviral construct expression. To determine the effect of gene activation on cell growth, 6×107 cells were sampled on days 7, 10, and 12 post-transduction. To compare the distribution of gRNAs in the transduced library with that in the original plasmid library, as well as between different virus preparations, 6×107 cells were sampled on day 7 post-transduction with either virus preparation. Although one might expect to see clusters of interacting cells in the library culture caused by the interaction of upregulated receptors, we did not observe any increase in cell aggregation, possibly due to the shear forces produced from this cell line being grown with constant shaking .
How A Ligand Works
The ligand travels through the watery fluids of an organism, within the blood, tissues, or within a cell itself. The ligand travels at random, but once the concentration is high enough, a ligand will eventually reach a protein. Proteins receiving ligands can be receptors, channels, and can even be the start of a complex series of intertwined proteins. When the ligand binds to the protein, it undergoes a conformational change. This means that while no chemical bonds have been formed or broken, the physical action of the ligand fitting into the protein changes the overall shape of the entire structure. This can trigger many actions. In most cases, the movement of the protein itself activates another chemical pathway, or triggers the release of another messenger ligand, to carry the message to other receptors.
It is this ability of the ligand, to activate a protein for a short amount of time and then be recycled, which allows for the biological control of many interactions. The amount of time a ligand spends attached to its receptor or specific protein is a function of the affinity between the ligand and the protein. If there is a high affinity, the ligand will tend to stick to the protein and modify its function for longer. If the ligand has a low affinity for the protein, it will be less likely to bond in the first place and will release from the receptor faster.
Recommended Reading: What Is Locus In Biology
Ligands Definition Types Function And Examples
Ligands: Ligands are those molecules or ions in a coordination compound that donate a pair of electrons to the central atom of the compound. Ligands can be positively charged, negatively charged or neutral metal atoms and help in binding the molecules together. The term Ligand is an English word derived from a Latin word Ligandus which means to bind. In most cases the ligands are found to form a covalent bond with the central atom.
Some examples of ligands are NO+, N2H5+ , Br, I, S2 or H2O, NO, CO . The ligands are classified on the basis of number of binding sites, size of the binding atom and the charge they carry. the concept of ligands is very crucial to understand the important chemical reactions studied in Inorganic and Organic chemistry. The students can find the complete details of the same in this article. Continue reading.
Simple Biochemical Networks Allow Accurate Sensing Of Multiple Ligands With A Single Receptor
-
Affiliations Department of Physics, Emory University, Atlanta, Georgia, United States of America, Computational Neuroscience Initiative, University of Pennsylvania, Philadelphia, Pennsylvania, United States of America
-
* E-mail:
Affiliations Department of Physics, Emory University, Atlanta, Georgia, United States of America, Department of Biology, Emory University, Atlanta, Georgia, United States of America, Initiative in Theory and Modeling of Living Systems, Emory University, Atlanta, Georgia, United States of America
Don’t Miss: What Is Spatial Science In Geography
The Recognition Of Unrelated Ligands By Identical Proteins
ACS Chem. Biol.
Publication History
Article Views are the COUNTER-compliant sum of full text article downloads since November 2008 across all institutions and individuals. These metrics are regularly updated to reflect usage leading up to the last few days.
Citations are the number of other articles citing this article, calculated by Crossref and updated daily. Find more information about Crossref citation counts.
The Altmetric Attention Score is a quantitative measure of the attention that a research article has received online. Clicking on the donut icon will load a page at altmetric.com with additional details about the score and the social media presence for the given article. Find more information on the Altmetric Attention Score and how the score is calculated.
Receptor Activator Of Nuclear Factor Kappa Beta Ligand
RANKL is a transmembrane molecule expressed by mesenchymal cell and lymphocytes. The soluble form of RANKL is a consequence of proteolytic cleavage. RANKL binds to RANK on hematopoietic cells and activates cytoplasmic adaptor proteins . These events activate downstream signaling pathways involving NF-kB, MAPK family, and NFATc1. However, RANKL is neutralized by its soluble decoy receptor OPG. It is consequently the ratio between RANKL and OPG that regulates the process of bone resorption. Based on this physiological principle, the monoclonal antibody that neutralizes RANKL may have efficacy to prevent fractures in osteoporotic patients. As a consequence of the importance of RANK/RANKL, this topic will be discussed further under bone resorption .
Fig. 6. Receptor activator of NF-kappaB ligand , RANK and osteoprotegerin build a triumvirate of molecules that control the formation, activity and the survival of osteoclasts. RANKL binds to RANK and activates cytoplasmic adaptor proteins . Downstream signaling pathways include NF-kB, MAPK family, and NFATc1. RANKL is neutralized by its soluble decoy receptor OPG. Thus, the ratio between RANKL and OPG regulates the process of bone resorption.
Prasoon Pandey, Neelam Balekar, in, 2018
Also Check: What Do You Learn In Physics Class
Classification Of Ligands As L And X
Especially in the area of organometallic chemistry, ligands are classified as L and X . The classification scheme the “CBC Method” for Covalent Bond Classification was popularized by M.L.H. Green and “is based on the notion that there are three basic types … represented by the symbols L, X, and Z, which correspond respectively to 2-electron, 1-electron and 0-electron neutral ligands.” Another type of ligand worthy of consideration is the LX ligand which as expected from the used conventional representation will donate three electrons if NVE required. Example is alkoxy ligands. L ligands are derived from charge-neutral precursors and are represented by amines, phosphines, CO, N2, and alkenes. X ligands typically are derived from anionic precursors such as chloride but includes ligands where salts of anion do not really exist such as hydride and alkyl. Thus, the complex IrCl2 is classified as an MXL3 complex, since CO and the two PPh3 ligands are classified as Ls. The oxidative addition of H2 to IrCl2 gives an 18e ML3X3 product, IrClH22. EDTA4 is classified as an L2X4 ligand, as it features four anions and two neutral donor sites. Cp is classified as an L2X ligand.
Differentiate Between Different Types Of Signals
There are two kinds of communication in the world of living cells. Communication between cells is called intercellular signaling, and communication within a cell is called intracellular signaling. An easy way to remember the distinction is by understanding the Latin origin of the prefixes: inter means between and intra means inside .
Chemical signals are released by signaling cells in the form of small, usually volatile or soluble molecules called ligands. A ligand is a molecule that binds another specific molecule, in some cases, delivering a signal in the process. Ligands can thus be thought of as signaling molecules. Ligands interact with proteins in target cells, which are cells that are affected by chemical signals these proteins are also called receptors. Ligands and receptors exist in several varieties however, a specific ligand will have a specific receptor that typically binds only that ligand.
Recommended Reading: How Does Australia’s Geography Affect Its Economy
Comparison Of Dcas9 Activators With P300 And Vp64
5×106 cells were transduced at < 0.3 MOI, with lentivirus-packaged gRNA pools, with each pool of 8 gRNAs targeting one of 12 surface proteins. This was to mimic screening conditions as closely as possible and to avoid synergistic activation caused by the expression of multiple gRNAs targeting the same gene in one cell. Transduction was carried out by incubating viral supernatants with cells at 37°C overnight . The next day, cells were reverse-transfected with respective dCas9-activator expression constructs using Lipofectamine LTX and PLUS Reagent in a 96-well format and grown for an additional 48h before analysis by antibody staining and flow cytometry. Transfection efficiency was estimated to be between 70 and 80% from analysis of cells transfected with the non-activating control expressing dCas9-eGFP .
Kinetic Proofreading For Approximate Estimation
The approximate solution can be computed by cells using the well-known kinetic proofreading mechanism . In the simplest model of KPR , intermediate states between an inactive and an active state of a receptor delay the activation. Thus bound ligands can dissociate before the receptor activates, at which point it quickly reverts to the inactive state. Since rc< rnc, cognate ligands dominate among bindings that persist to activation. The resulting increase in specificity in various KPR schemes has led to their exploration in the context of detection of rare ligands . Instead, here we analyze their ability to measure concentrations of both ligands simultaneously. We first consider the case where both the cognate and the non-cognate ligand concentration are comparable, cc cnc and the dissociation rates are distinct, rc rnc. In the following sections, we explore another case, cc cnc and rc rnc, a situation common in immunology.
Consider a biochemical network in Fig 4: the receptor, R, activates two messenger molecules, A and B. The former is activated with the rate kA only if the receptor stays bound for longer than a certain Tc . The latter is activated with the rate kB whenever the receptor is bound. The molecules deactivate with the rates rA and rB, respectively, and all activations/deactivations are first-order reactions. Then the mean concentrations of the messenger molecules are :
You May Like: Who Got The First Nobel Prize In Physics
Antiprogrammed Cell Death Receptor Ligand 1
PD-L1 is the ligand for PD-1, and ligation of PD-L1 to PD-1 leads to T-cell inhibition.102 PD-L1 is expressed in a multitude of tissues including muscles and nerves. Of relevance for cancer immunotherapy, PD-L1 can be expressed on the surface of tumor cells, tumor-associated macrophages , and T lymphocytes and can subsequently inhibit PD-1positive T cells.103,104 The expression of PD-L1 can be induced by cytokines such as IFNs, or alternatively PD-L1 can be expressed autonomously through aberrations in the EGFR signaling pathway.105108
In clinical studies, antibodies blocking the PD-L1 and PD-1 interactions have demonstrated 621% ORR in non-selected tumors,109110 19% in triple-negative breast cancer,111 and 38% in NSCLC demonstrating high PD-L1 expression.112 Zandberg and colleagues114 showed slightly less efficacy than the PD-1 inhibitors, with a preliminary ORR of 11%, 71% of which were durable at 55 weeks and somewhat dependent on PD-L1 expression. The toxicity profile for antiPD-L1, like that of antiPD-1, is thought to be distinct and generally milder compared with the adverse events observed with antiCTLA-4.110
The mechanism of action of antiPD-L1 is currently under investigation. However, several studies have shown that clinical benefit is directly correlated with high expression of PD-L1.112
David R. Calabrese, … John S. SchneeklothJr., in, 2019
Spectrochemical Series Of Ligands
Crystal field splitting occurs due to the field produced by ligands on the central metal atom. Thus a series of absorption spectra caused due to absorption of light as a result of this splitting of energy levels is known as spectrochemical series.
Based on the result of absorption spectra of cobalt complexes, a spectrochemical series was found in \ It is a list of ligands arranged in a series according to their field strength, i.e. based on the splitting of energy levels. Based on the splitting magnitude, ligands are classified into two types:
1. Strong Field Ligands Ligands that cause large splitting of energy levels. They form low spin complexes. For example \ etc.
2. Weak Field Ligands Ligands that cause less or small splitting of energy levels. They form low spin complexes. For example \}\left,\) etc.
Given below the spectrochemical series of ligands:
Read Also: What Do We Study In Psychology
Types Of Liagands On The Basis Of Denticity
If the ligand is attached to the central metal atom only by one coordinate bond, then the ligand is called a monodentate or unidentate ligand. For example, unidentate ligands are Cl, NO2, Br, CN, OH H2O, NH3, CO, etc.
If ligands having two or more donor atoms are called polydentate ligands. They are further classified as bidentate , tridentate , tetradentate and those with five and six atoms are called Penta and hexadentate ligands respectively.
For example, In 4-, CN is a monodentate ligand. In ethylenediamine, , a neutral ligand, is a bidentate ligand in ethylenediamine copper complexion. CO32-, SO42-, 2- are bidentate ligands with a double negative charge.
Briefly, we can say that monodentate, bidentate, tridentate, tetradentate, pentadentate, hexadentate have one, two, three, five, and six donor sites respectively.
Bmp Receptor Activity In The Ovary
The BMP ligands strongly activate the BMPR1B, and their role in the regulation of gonadotrophin receptor expression has been previously reported . The BMPR1B is first expressed in primordial follicles on the oocyte and the granulosa cells of primary follicles throughout folliculogenesis . Androgen receptors are first expressed in the transitional follicle between the primordial and primary stage, and are early regulators of ovarian development, particularly the inducement of FSHR on the granulosa cell .
Fig. 6. The stage-specific relationship between granulosal receptor expression and estrogen activity during folliculogenesis in a natural cycle. Dominant follicle selection took place when the androgen receptor and FSHR expression decreased, and LHR expression increased . Downregulation of FSHRs, LHRs, and the cessation of proliferation occurs in the preovulatory follicles in humans and animals . CYP19a1 is the gene that encodes aromatase, essential for the production of estrogen.
Fig. 7. The stage-specific relationship between granulosal receptor expression during folliculogenesis in human IVF cycles. Granulosal BMPR1B, FSHR, and LHR protein density and follicle size profile of young patients with a typical ovarian reserve for the age group. The patients were 2330 years old and stimulated with gonadotrophins during an IVF cycle. Values in graphs are means ± SEM, and differences were considered significant if *P& lt 0.05 .
Animal Reproduction Science, 111,
Don’t Miss: Basis And Span Linear Algebra