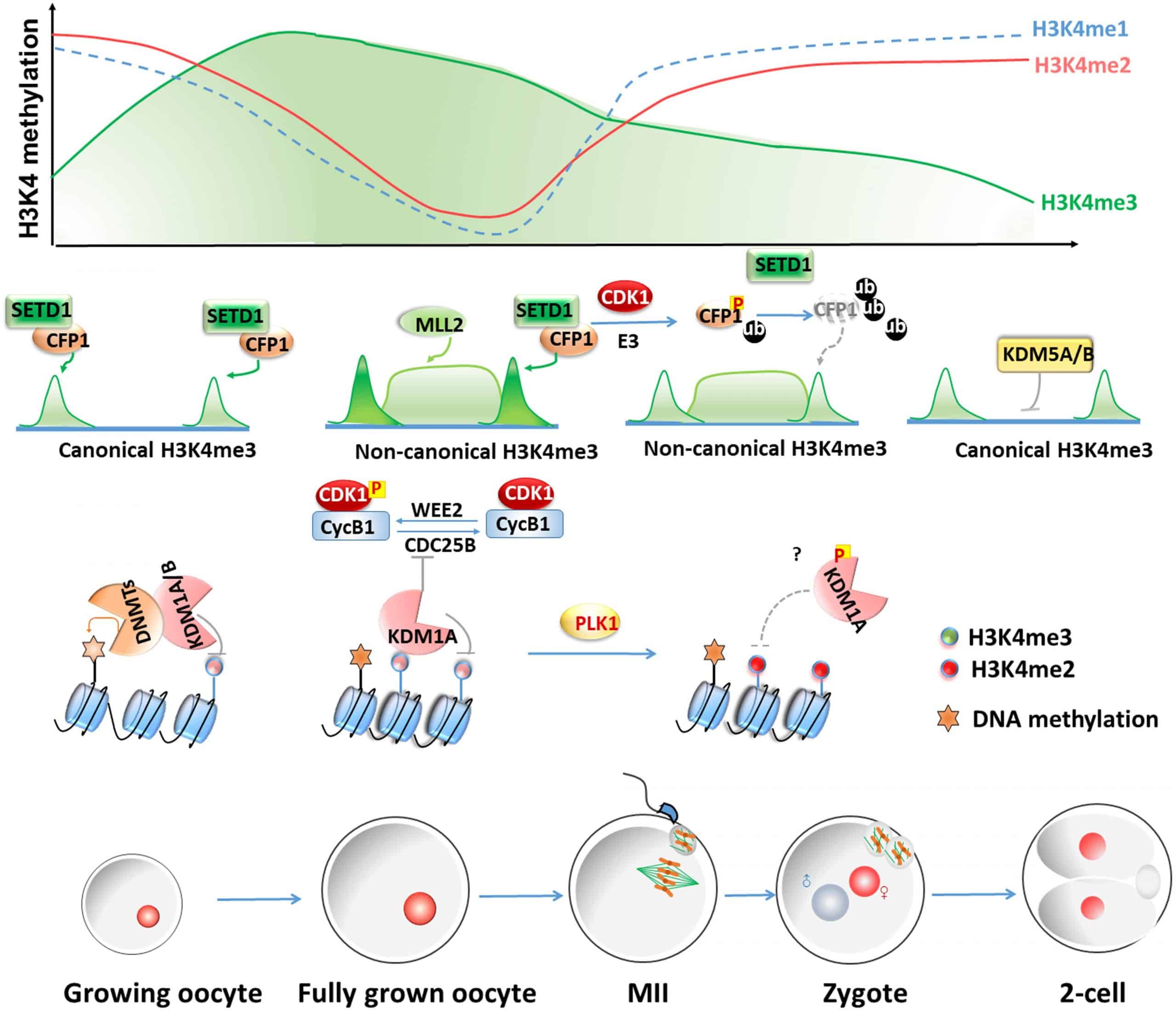
Dynamics Of Dna Methylation
Variation in DNA methylation has been detected in many organisms, including viruses, prokaryotes, and eukaryotes . Methylation of DNA plays important roles in the regulation of gene expression, growth, development, and protection from environmental stresses, as well as in stabilizing the genome . DNA base modification in a context-and genomic region-specific manner is catalyzed by different enzymes through distinct pathways. Methylcytosine , also known as the fifth base of DNA, was discovered long before the DNA was recognized as genetic material in a living cell. Although more attention is given to the conventional 5-mC, recent findings on additional base modifications have resulted in overwhelming interest in epigenomic studies. In plants, cytosine methylation can occur in all contexts of cytosine . In Arabidopsis as well as in other plants, the heterochromatic regions are enriched with methylcytosines, generally in the repetitive sequences and TEs. However, TEs and 5-mC are also found to be interspersed in the euchromatic regions . The dynamics of DNA base methylation depends on the reversibility of the processes, which also controls switching on/off the gene. Diversity and complexity of epigenetic changes in different organisms are being discovered continuously, and the potential combinatorial interactions of epimarks indicate that epigenetic codons would be considerably more complex than it is thought today .
Essential Steps To Take Prior To Addressing Methylation :
- Know your Homocysteine level this is a blood test that can be done at a regular lab.
- Know your methylation SNPs order a 23andme saliva test kit, then when the results are in, give the data to your methylation practitioner who will run it through software that identifies your SNPs.
- Check your urine sulfur level your methylation practitioner will be able to guide you on how to do this.
- Complete a trial of taking hydroxo-B12 for at least 5 days your methylation practitioner will guide you.
Once you have completed these steps, and if your homocysteine level is higher then 7, then your methylation practitioner will guide you to start taking methyl-folate, along with other important B vitamins in the methylation cycle, starting with low dosages. Each persons body responds differently as the methylation cycle optimizes, so it is important to go slowly so that we can find out how your body will respond and address any adjustments that need to be made.
This is how your methylation cycle gets the nutrients it needs to keep you healthy. And it is NOT just about methyl-folate. The next steps in methylation cycle need to be addressed as well. Methyl-folate connects with the next dominos in the chainMTR, MTRR, MAT, BHMT, and finally CBS.
BHMT is a shortcut through the middle of the methylation cycle that allows your body to use choline instead of folate and B12 to make methionine, which is turned into SAM by MAT.
Methylation C677t Half Of Us Have It
About 45 percent of people have this genetic trait, what is referred to as a MTHFR defect, so feel free to blame mom and dad for that. Thats about 1 in 2 of people that have 1 copy of the C677T MTHFR gene . But its not just about genes. Disturbances in this pathway can occur for other reasons, I am pretty sure you are going to find yourself in the next list.
Recommended Reading: What Was The Most Significant Error Of Ptolemy’s Geography
Dna Methylation And Its Machinery
DNA methylation is a process by which methyl groups are added to the DNA molecule, especially at the 5 carbon of the cytosine ring, which forms 5-methylcytosine . In mammals, 5mC is mostly found in the context of paired symmetrical methylation of a CpG site, a site in which a cytosine is located next to a guanidine . However, non-CpG methylation is also detected in human and other species . In the bulk of genomic DNA, most CpGs are methylated, whereas those located in a CpG island remain largely unmethylated .
DNA methylation is mediated by DNMTs. In mammals, five family members of the DNMT proteins have been characterizedDnmt1, –2, –3a, –3b, and –3Lyet only the first three possess DNMT activity . DNMT1 is the maintenance Dnmt for replication, whereas DNMT3a and -3b are referred as de novo DNMTs, as they can establish a new DNA methylation pattern . The DNMT3-like protein Dnmt3L is homologous to the other Dnmt3s but lacks catalytic activity, and Dnmt2 has sequence homology to all Dnmts but methylates cytoplasmic tRNA instead of DNA .
Dna Methylation In The Developing Cns
The precise temporal regulation of de novo methylation and demethylation is particularly important for the differentiation and maturation of the mammalian central nervous system . Multipotent neural progenitor cells sequentially undergo neurogenesis and astrogliogenesis . In particular, the differentiation switch of NPCs from neurogenesis to astrogliogenesis coincides with DNA methylation and demethylation events on the glial fibrillary acidic protein gene promoter region . Early in neurogenesis at E11.5, DNA methylation of the Gfap promoter represses its expression . The continual expression of Dnmt1 in NPCs has been found to be important for the maintenance of the methylation pattern on the Gfap promoter through subsequent cell divisions . Interestingly, neurogenesis from E11.5 to E14.5 is the only time during neural development that Dnmt3b is strongly expressed before declining to nearly undetectable levels in the CNS . At E14.5, the Gfap promoter undergoes DNA demethylation to coincide with the differentiation of the astrocytic lineage . As development further progresses, the decline of Dnmt3b and the peak expression of Dnmt3a at 3 weeks postnatally coincide with remethylation and reduced transcription of the Gfap promoter . The coordinated expression of Dnmts and their ability to regulate the methylation pattern of the Gfap promoter organize and regulate neuronal development.
You May Like: What Is Relief In Geography
Canonical Rddm And Non
The second step of canonical RdDM is Pol V-mediated de novo methylation. Here, Pol V is recruited to target loci by the SU3-9 homologs SUVH2 and SUVH9, which bind to methylated DNA . Pol V then transcribes a class of long non-coding RNAs, thought to serve as scaffolds for RdDM targeting . During this step, either AGO4 or AGO6 binds 24-nt siRNAs and are recruited to Pol V by interaction with the largest subunit of Pol V and KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR 1 . The 24-nt siRNAs can then pair with nascent scaffold RNAs produced by Pol V . AGO4/AGO6 then recruits DOMAINS REARRANGED METHYLTRANSFERASE 2 , a homolog of mammalian DNMT3, to catalyze DNA methylation. It is thought that RNA-DIRECTED DNA METHYLATION 1 may assist in the AGO4DRM2 interaction, however, its role remains unclear .
Many of the proteins involved in RdDM arose through gene duplication, and subsequent sub and neofunctionalization . Some key components have only been found in angiosperms . For instance, Pol IV and V evolved from duplication of various subunits of Pol II , and there is evidence of further lineage-specific duplication and evolution of a putative Pol VI in grasses . From an evolutionary perspective, RdDM is a powerful example of how novelty and complexity can arise through gene duplication.
Basic Mechanism Of Dna Methylation
The enzymes that establish, recognize, and remove DNA methylation are broken into three classes: writers, erasers, and readers. Writers are the enzymes that catalyze the addition of methyl groups onto cytosine residues. Erasers modify and remove the methyl group. Readers recognize and bind to methyl groups to ultimately influence gene expression. Thanks to the many years of research devoted to understanding how the epigenetic landscape is erased and reshaped during embryonic development, many of the proteins and mechanisms involved in DNA methylation have already been identified.
Don’t Miss: Is Clinical Psychology In Demand
Dna Methylation In The Etiology Of Neurological And Psychiatric Disorders
The pattern of DNA methylation established during development can be modulated by neural activity in order to encode learning and memory. When the mechanisms that establish and recognize the DNA methylation pattern are dysfunctional, problems with learning and memory frequently result. One of the most common forms of mental retardation, Rett Syndrome, is frequently caused by a mutation to the methyl-binding protein MeCP2 . The onset of symptoms at 618 months of age coincides with a time in early development when sensory experience is driving dendritic pruning and shaping connections in the brain . Although MeCP2 is expressed by the majority of cells, it is particularly important for normal neuronal function. In mice, loss of MeCP2 in neurons is sufficient to recapitulate the majority of Rett symptoms . The phenotype of MeCP2 mutant mice can be reversed by restoration of the MeCP2 gene in postmitotic neurons . As previously mentioned, MeCP2 is regulated by neuronal activity and in turn regulates the expression of BDNF, which has enhanced expression following depolarization . The overexpression of BDNF in postmitotic neurons of MeCP2 mutant mice ameliorates their phenotype, suggesting that MeCP2 is critical for regulating the expression of genes like BDNF that are regulated by neuronal activity and essential for normal cognitive function . The role of MeCP2 in Rett Syndrome will be further discussed in later chapters.
Ink4a Methylation In Bladder Cancer
Inactivation of theINK4A gene may occur by a variety of mechanisms including deletion, mutation, and promoter methylation . Mutation or deletion of oneINK4A allele and concurrent methylation of the remaining allele results in complete loss of functional activity. Methylation ofINK4A has been reported in 27% to 60% of primary urothelial carcinomas . Such epigenetic changes are among the earliest molecular events associated with transformation and may therefore precede morphologic alterations in cellular architecture.
The first detailed study investigating the effects ofINK4A promoter methylation on transcriptional activity was performed in bladder cancer cells, where a reduction inINK4A expression was associated with higher levels of methylation in the upstream promoter region methylation of specific CpG sites in theINK4A promoter was shown to significantly downregulate transcriptional activity of the gene . Administration of the demethylating agent 5-aza-2-deoxycytidine was capable of reactivatingINK4A expression in bladder cancer cells that were previously shown to contain methylated alleles of the gene. Methylation in exon 2 of theINK4A gene is also a frequent occurrence in a variety of cancers and is an excellent marker for transformation, although the presence of methylation in this region of theINK4A gene does not affectINK4A transcription in bladder cancer cells .
Sophie J. Deharvengt PhD, Gregory J. Tsongalis PhD, in, 2018
Also Check: Unit 1 Geometry Basics Homework 2 Segment Addition Postulate
Understanding The Methylation Cycle And Its Effect On Health
Part 3 of Dr. Donis Series on How Genetic Mutations Affect Your Health
In the first two parts of this blog series, we explored how genetic mutations can affect our health and how we go about finding out which SNPs you have as the first step on the road to optimal health.
Understanding your genetic mutations will help you identify which processes and enzymes may need support in your body and your metabolism.
Once you have this information youll be able to design exactly the right support for your body including making the right diet choices, taking the right nutrients and optimizing the way in which you respond to stress, all based on your individual needs.
Today Im going to discuss the enzymatic pathways in the methylation cycle that influence not only the way you feel from day to day but also your risk of disease in the long run. Then, next week, I will share my 8 steps to a healthy methylation cycle. First, before diving into the enzymes, lets talk about why the methylation cycle is important and how it affects your health.
Repression Of Transposable Elements
DNA methylation is a powerful transcriptional repressor, at least in CpG dense contexts. Transcriptional repression of protein-coding genes appears essentially limited to very specific classes of genes that need to be silent permanently and in almost all tissues. While DNA methylation does not have the flexibility required for the fine-tuning of gene regulation, its stability is perfect to ensure the permanent silencing of transposable elements. Transposon control is one of the most ancient functions of DNA methylation that is shared by animals, plants and multiple protists. It is even suggested that DNA methylation evolved precisely for this purpose.
Don’t Miss: 5.3 Exercises Algebra 2
Dna Methylation And Transcription Factor Binding Dynamics
Furthermore, it is worth mentioning that enzymatic modification of cytosine is a complex dynamic involving DNMT1, DNMT3A and DNMT3B methyltrasferases, which methylates cytosines , and the TET family of cytosine oxygenase enzymes, which oxidizes 5mC to 5-hydroxymethylcytosine , subsequently to 5-formylcytosine and finally to 5-carboxycytosine . These oxidized derivatives might also hinder TF binding. In principle, the presence of these derivatives can alter the way in which proteins bind to their recognition sequences in DNA by strengthening the interactions, weakening them, or by abolishing them completely. For example, Klf4 shows the strongest binding to fully methylated DNA, with slightly higher affinity than that of the unmodified DNA, and in each oxidation event, from 5mC to 5hmC to 5fC to 5caC, resulting in progressively weaker binding .
The Gut Is Also In Charge Of Methylation
High-quality probiotics are incredibly important to people with a methylation problem because if you let Candida overrun your gut, you get excessive amounts of Candidas toxin called acetylaldehyde. I should tell you that acetylaldehyde is also a break down product of drinking alcohol. So job one is to repair the digestive tract and stop drinking alcohol. Optimize gut flora. The less candida you have, the less acetylaldehyde. You may have yeast overgrowth and not even know it. People who have been drinking a long time have been mugged of Thiamine and probiotics. Read my Drug Muggers book for ways to correct that.
Here are the symptoms of a HANGOVER, as well as YEAST overgrowth:
- Tender points or soreness
- These symptoms also happen in people who have reduced methylation!
If youve taken an antibiotic for more than a week, you are low in probiotics. If you have had your appendix removed, you are deficient in probiotics . If you drink a lot of coffee, if you have recurrent vaginal yeast infections, if you have a lot of flatulence, if you crave sweets or have a white coating on your tongue you are deficient in probiotics.
Ive negotiated a coupon code suzy12 from the owner of this website which gives you free shipping via US mail, and a generously sized sample beauty bar of Kampuku think of it like probiotic soap, its amazing for skin conditions. I personally use this, and love it. Now, back to how to this yeast story.
You May Like: Who Is Ronan Farrows Biological Father
Methylation Quantitative Trait Loci
Given the observed evidence for DNA methylation heritability, much interest has focused on identifying specific genetic variants that influence DNA methylation variation across the genome. Multiple studies have explored the correlation between DNA methylation levels and genetic variants across the genome , to identify DNA methylation quantitative trait loci or meQTLs . Although several early papers tackled meQTLs identification over limited target sites , it was not until the early 2010s that initial genome-wide efforts identified meQTLs on the 27K methylome and across multiple tissues .
Studies to date have reported an influence of meQTLs on up to 45% of CpG sites profiled by the Illumina 450K array across the genome , with more than 90% of meQTLs acting on nearby methylation sites . CpG sites that have higher heritability estimates are more likely to be associated with meQTLs in cis, trans, or both, and have a clear polygenic architecture . Some studies also include replication in independent sample sets, although overall a direct comparison of meQTL signals can be challenging because studies do not systematically report meQTL effect sizes. Despite observations that meQTLs tend to have moderate to large effects, the missing heritability issue has also been raised in the context of meQTLs. That is, family-based heritability estimates of DNA methylation are greater than the proportion of variance explained by meQTLs, especially for distal associations .
What Is Meant By The Term Methylation
The term methylation is used to refer to the addition of a methyl group to a given reactant. It can also refer to the substitution of one of the functional groups on a given reactant with a methyl group. The process of methylation can be thought of as a type of alkylation in which a methyl group is interchanged with a hydrogen atom on a substrate.
You May Like: Why Is Chemistry Called The Central Science
Milestones In The Development Of Targeted Dna Demethylation Tools
In comparison with targeted DNA methylation, tools to induce targeted DNA demethylation have a shorter history, probably in part because there is no single mechanism to remove methylation directly in mammals. Active removal of 5mC demethylation involves iterative oxidation and thus requires multiple steps. In one pioneering study, Gregory et al. selected the Rel-homology domain , a well-characterized NFkB-binding domain, to anchor thymine DNA glycosylase via a short glycine-rich linker. These authors observed a loss of DNA methylation at the targeted locus as well as increased transcription of Nos2 in the NIH3T3 cell line. In order to increase the flexibility of targeting, the group replaced RHD with ZFs. Using whole-genome expression microarrays and pathway analysis, the authors found that the targeted demethylation of Nos2 affected only 42 genes, and that the majority of these genes were downstream of Nos2 . These studies demonstrate that targeted DNA demethylation using TDG can upregulate gene expression.
Fig. 3
Eukaryotic Deoxyribonucleic Acid Methylation Involves Two Dynamically Regulated Metabolic Pathways
Several DNA methylation processes are observed in cells: de novo cytosine methylation, maintenance methylation during replication of double-stranded DNA , active demethylation during the absence of replication, and spontaneous demethylation when maintenance methylation is suppressed. CpG sites are the primary sites of cytosine methylation in eukaryotic DNA, but methylation of other than CpG sites occurs.
Figure 3. DNA maintenance methylation in eukaryotes. The chief function of DNA maintenance methylation in eukaryotes is the faithful transmission of the pattern of methylation from generation to generation. Replication of symmetrically methylated DNA results in hemimethylated DNA followed by maintenance methylation catalyzed by DNA methyltransferases, methylate unmethylated cytosines in CG or CXG motifs to restore the original pattern of symmetrically methylated DNA.
Don’t Miss: How Much Do Math Teachers Make