Split Genes Or Interrupted Genes
Let us make an in-depth study of the split genes or interrupted genes. After reading this article you will learn about: 1. Processing of mRNA by Splicing 2. Spliceosome 3. Mechanism of Splicing 4. Self Splicing and 5. Alternate Processing of Pre-mRNA.
Processing of mRNA by Splicing:
Till recently it was believed that coding sequence of DNA and amino acids of polypeptide is collinear. The coding sequences are continuous and codon for one amino acid is adjacent to the codon for the next amino acid. The open reading frame is a single stretch of codons without any gap.
But now it has been discovered that coding sequence of most of enkaryotic genes is split into stretches of codons interrupted by stretches of non-coding sequences. Most human genes are discontinuous.
The coding sequences of DNA of the gene are called exons. In between exons, there are intervening non-coding sequences called introns. This type of genes are called split genes or interrupted genes. They are most common in enkaryotes. They are also found in viruses but rarely in bacteria.
The terms exons and intons were given by Gilbert in 1977. It was discovered in Amphibia, mammals and some other animals that genes are not represented by continuous sequence of nucleotides. Introns are removed by excision and discarded.
Translation takes place only after the splicing is completed.
RNA Splicing:
Spliceosome:
Mechanism of Splicing:
Self Splicing:
Alternate Processing of Pre-mRNA:
1. Alternate Splicing:
The Central Dogma Of Protein Synthesis
We know that an organisms genetic information is stored in its genes, the functional subunits of the genome. Genes are arranged in the strands of the DNA double helix in the nucleus of cells.
We also know that this information is transcribed from DNA into a messenger RNA template in a process called transcription. As the name suggests, this mRNA template then acts as a messenger to synthesize proteins in a process called translation.
This is the first part of what is generally known as the central dogma of molecular biology, which was first described by Francis Crick in 1958.
But, as you probably know by now, biology is never that simple. After uncovering the general mechanisms behind protein synthesis, it was later discovered that in viruses, some non-coding mRNA coding sequences, known as introns, need to be removed or spliced out from the final mRNA molecule before it can be translated into protein.
Removal of these introns leaves only the protein-coding regions, called exons, which must be joined by RNA splicing to produce mature mRNA to allow for the translation of a functional protein. These findings were later extended to other organisms including eukaryotes.
As a quick summarymost genes in higher eukaryotes are transcribed as pre-mRNA, which contains non-coding and coding regions known as introns and exons, respectively.
In a process mediated by the spliceosome, introns are removed while exons remain to give a final mature mRNA sequence.
Recognition Of The Rna Elements
The conserved sequence motifs of the pre-mRNA are mainly recognized by conserved RNA elements from both snRNA and pre-mRNA. Three nucleotides at the 3-end of the 5-exon are anchored on the loop I of U5 snRNA in the Bact through P complexes. The BPS and surrounding sequences of the intron form an elongated duplex with complementary sequences of the U2 snRNA in the A through ILS complexes. The dinucleotides GU at the 5 end of the 5SS are recognized by the Linker domain of Prp8 in the S. cerevisiae U4/U6.U5 tri-snRNP and the human B complex and by Cwc24 in the Bact complex the six consecutive nucleotides downstream from GU of the 5SS are recognized by U6 snRNA in the Bact through ILS complexes. In the P and possibly C* complexes, the dinucleotides AG of the 3SS are recognized by the lariat junction that is formed after the branching reaction . Notably, AG of the 3SS pair up with the nucleophile-containing A of the BPS and the invariant G at the 5-end of the 5SS, respectively, through noncanonical WatsonCrick H-bonds.
You May Like: Geography And Geology Difference
Putative Mechanisms Of Trans
Currently, the mechanisms underlying trans-splicing in vertebrates remain largely unknown. Little is known about how the associated partner genes are physically recruited and what factors are involved in the process. Based on previous studies, we summarize several current models and propose new ones to address these issues.
The Assembly Of The Enzyme Substrate Complex
The spliceosome, like many macromolecular machines, is not pre-assembled as an active enzyme. A simple enzyme must, as any catalyst, be unchanged at the end of the reaction it catalyzes.20 The spliceosome is an enzyme in pieces, and thus to fit the strict definition of an enzyme, these pieces must be regenerated so that they may catalyze multiple rounds of pre-mRNA splicing. A consideration of the number of introns spliced and the maximum number of spliceosomes potentially active at any one time leads to the conclusion that spliceosomal components are involved in multiple splicing reactions.9,14 The whole system involved in the assembly of the spliceosome on a pre-mRNA and in the regeneration of active pieces 6 can, therefore, be considered to be an enzyme.
The in vitro pathway of enzyme-substrate assembly can be diagrammed as follows
The splicing substrate , U1 snRNP , protein factors , the commitment complex , U2 snRNP , the pre-spliceosome or complex A , U5 · U4/U6 tri-snRNP complex , the immature spliceosome or complex B , and the spliceosome or complex C are indicated. The enzyme is designated as E.
Matthew L. Kahlscheuer, … Nils G. Walter, in, 2015
Also Check: Hawkes Learning Systems Answer Key
Splicing And Human Disease
Another area where progress is particularly evident is in understanding the role of splicing in human disease and in applying this understanding to new therapeutic approaches. Tito Baralle described how the effect of mutations in splicing regulatory elements is dependent on genetic background. His team has found that because alternative exons are frequently controlled by multiple elements, mutations in one regulatory sequence may be silent on their own, but can make an exon more dependent on other elements. The complexity of this interplay was further highlighted by Cyril Bourgeois in regard to splicing of dystrophin exon 31 and by Joerg Gromoll for luteinizing hormone receptor exon 10. In each case, exonic mutations destroy or create binding sites for splicing regulators that alter the splicing of the exon and cause human disease.
The range of questions being addressed and the variety of techniques described at the meeting made clear that the field of alternative splicing studies is robust and growing . Important progress has been made in a multitude of directions, but the combination of genome-wide analyses with focused genetic or biochemical assays is proving to be particularly powerful. The meeting spurred very useful dialog between these global and more focused views, and we are looking forward to the next such gathering.
Intrinsic Disorder And Alternative Splicing
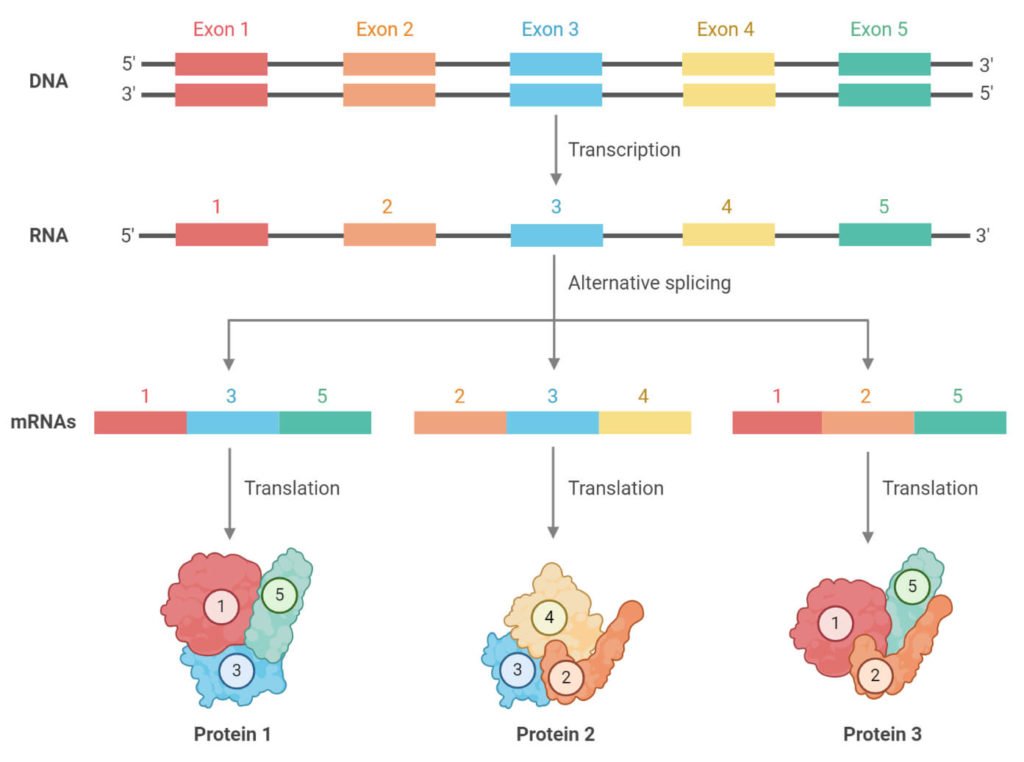
During RNA splicing, the exons of the pre-mRNA can be reshuffled and reconnected in multiple ways due to the alternative splicing . Due to this process, a single gene may code for multiple proteins and in humans, 95% of multiexonic genes are alternatively spliced and > 60% genes yield proteins via the AS mechanism. This provides an important mechanism for enhancing protein diversity in multicellular eukaryotes and affects functional diversity of proteins. Abnormal AS is associated with numerous human diseases, such as myotonic dystrophy, axoospermia, Alzheimers disease, Parkinsons disease, and cancer. In ordered proteins, regions affected by AS are relatively rare, small and are mostly found in coil regions located on the protein surface. However, AS is commonly found in IDPs, and disordered regions affected by AS are numerous and show very wide length distribution. Therefore, AS regions of mRNA code for intrinsically disordered regions much more often than for structured regions. This linkage between AS and signaling by disordered regions provides a novel and plausible mechanism that could underlie and support cell differentiation, which ultimately gave rise to multicellular organisms in nature.
Mario Blanco, Nils G. Walter, in, 2010
Also Check: Holt Geometry Lesson 4.5 Practice B Answers
What Is Mrna Splicing And Why Is It Important
mRNA splicing is a form of post-transcriptional modification of the mRNA transcript. DNA is made up of coding regions called Exons. These determine the amino acid sequence of a polypeptide chain during protein synthesis. However, it is also made up on non-coding regions called Introns that have no use when it comes to protein synthesis. Transcription of DNA produces pre-mRNA that has both Introns and Exons. To make mature mRNA the all Introns must be removed by a splicesosome made up of snRNA’s that recognize the Intron-Exon border these effectively ‘cut’ the Intron out and ligate the Exon ends together. This creates mature mRNA made up of just Exons. This is important because our DNA is made up of around 23,000 genes yet we have many more proteins than genes, so how is this possible? Splicing allows one gene to code for multiple proteins by altering which exons will remain in the mature mRNA transcript . This will change the sequence of amino acids in the primary structure of the protein which will in turn effect protein folding giving rise to proteins with different structures with different purposes from a single gene.
The Active Site Of The Spliceosome
Spliceosomes must bring together distant regions of the pre-mRNA along with spliceosomal snRNAs and proteins that enable catalysis. Alignment of the reactive groups occurs in part through scaffolding of the snRNA and pre-mRNA through the basepairing interactions between the intron and the U2 and U6 snRNAs ). These reactive groups must also be aligned with the U6 ISL, and it is thought that spliceosomal proteins such as Prp8 and the NTC play key roles in juxtaposing all of these functional groups. A number of experiments have found evidence that the U6 ISL plays a key role in coordinating essential magnesium ions for catalysis . Despite the necessity for proper alignment, the spliceosome active site must also be flexible since it is remodeled between the first and second steps of splicing to permit juxtaposition of the 5 exon and 3 splice site for exon ligation.
et al.
Mariano A. Garcia-blanco, … Sagarmoy Ghosh, in, 2001
You May Like: How To Find The Ksp
Conservation Of Splicing Machinery
We believe that trans-splicing shares most characteristics with cis-splicing. Several lines of evidence have shown that trans-splicing utilizes a similar set of splicing machinery to alternative splicing. Trans-splicing has the same splicing signals and factors as alternative splicing. For example, the spliceosome, which contains U1, U2, U4, U5, and U6 snRNAs, catalyzes pre-mRNA in cis-splicing. A recent study demonstrated that U1 snRNP binding may promote mod trans-splicing in Drosophila . For SL splicing, the key elements of SL snRNP are very similar to the spliceosomal snRNPs , indicating that SL RNA may originate from a splicing U snRNA in lower organisms with an ancestral cis-splicing mechanism. Additional support comes from the fact that SL trans-splicing exists in some metazoans, including cnidarians, ctenophores, rotifers, flatworms, nematodes, crustaceans, sponges, and chaetognaths. In contrast, plants, fungi, insects, most protists, and vertebrates do not exhibit SL trans-splicing . However, simultaneous trans-splicing events could take place between SL RNA and inherent transcripts in HeLa cells both in vivo and in vitro . In addition, SL trans-splicing is favored in adenosine-rich 5 -UTRs in hydrozoans . In vertebrates, the 5 -UTR can be involved in the generation of some trans-spliced mRNA chimeras , which is similar to SL trans-splicing in invertebrates.
Dna Methylation And Alternative Splicing In Social Insects
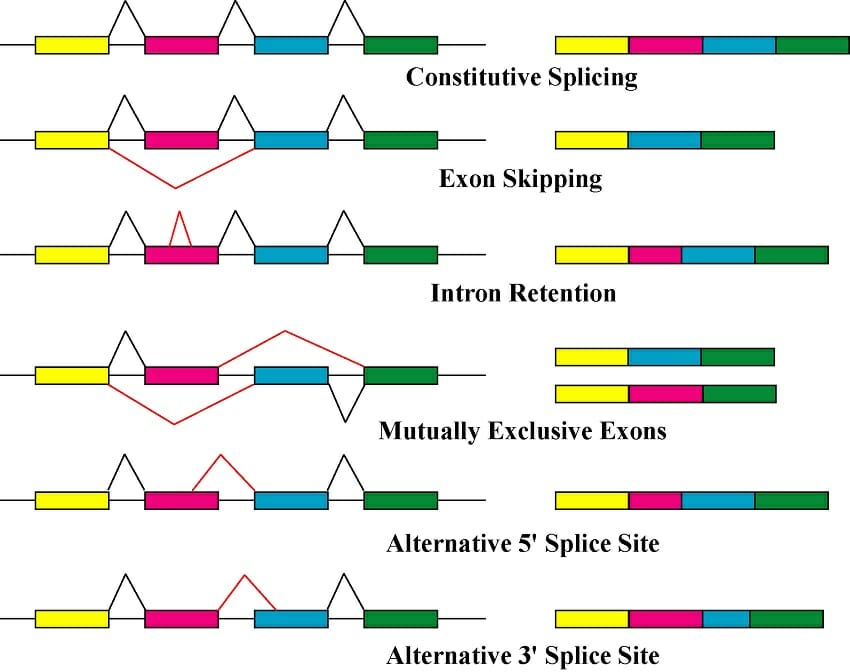
CpG DNA methylation has showed a role to regulate the alternative splicing in social insects. In honey bees , CpG DNA methylation seems to regulate the exon skipping based on the first few genomic studies after honey bee genome was available. CpG DNA methylation regulated alternative splicing more extensively, not only affect exon skipping, but also intron retention, and other splicing events.
Exon skipping: Drosophiladsx
dsx
Pre-mRNAs from the D. melanogaster gene dsx contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a polyadenylation signal in exon 4 causes cleavage of the mRNA at that point. The resulting mRNA is a transcriptional regulatory protein required for female development.
Alternative acceptor sites: DrosophilaTransformer
DrosophilaTransformer
Exon definition: Fas receptor
This mechanism is an example of exon definition in splicing. A spliceosome assembles on an intron, and the snRNP subunits fold the RNA so that the 5′ and 3′ ends of the intron are joined. However, recently studied examples such as this one show that there are also interactions between the ends of the exon. In this particular case, these exon definition interactions are necessary to allow the binding of core splicing factors prior to assembly of the spliceosomes on the two flanking introns.
Repressor-activator competition: HIV-1 tat exon 2
Also Check: Elastic Definition Physics
Challenges In Microarray Design For Splice Variant Detection
Microarray based gene splicing detection poses some unique challenges in designing probes for isoforms that show a high degree of homology. In order to differentiate between these isoforms, a microarray that uses a combination of probes for exons and exon-exon junctions is used. Exon skipping events or other deletions can be monitored by using junction probes. For example, a probe spanning the exon 1 and exon 3 of the gene will detect the skipping of exon 2 from the gene that is translated into a protein.
Cd4: An Example Of Alternative Splicing
For example, several alternatively spliced genes are known to be involved in immunity.
Studies indicate that alternative splicing of CD44, a protein involved in T-cell homing with 10 variable cassette exons and six distinct protein isoforms, is crucial for T-cell function.
The variable exons of CD44 encode portions of the membrane-proximal extracellular domain of the protein, and the presence of some of the variable exons has been shown to increase the association of CD44 with various proteins.
Isoform expression is activation dependent. This allows naïve T cells to express the smallest CD44 isoform, which lacks all variable exons.
In comparison, activated T cells express multiple CD44 isoforms, thereby suggesting that CD44 alternative splicing is important for activation.
Don’t Miss: Paraguay Geographical Features
How Do We Study Alternative Splicing
in vitroreverse transcription polymerase chain reaction7Human Proteome ProjectReferences
The Role Of The Protein Components
The protein components in each spliceosome can be divided into four distinct groups: the structural proteins, the splicing factors, the RNA-dependent ATPase/helicases, and other regulatory proteins. The structural proteins sustain the splicing active site conformation, support the overall characteristic appearance of the spliceosome, and provide the elasticity that is needed for the splicing reaction. The splicing factors facilitate assembly and activation of the spliceosome and assist the two steps of transesterification. The ATPase/helicases remodel the spliceosomal complexes to allow flux of the splicing factors and other proteins and RNA elements. The other regulatory proteins modulate the splicing of pre-mRNA and will be briefly discussed in a later section.
You May Like: The Founder Of Behaviorism Was:
Evolutionary Trends Of Trans
Trans-splicing frequently occurs in lower organisms, such as dinoflagellates , euglenozoa , and some species of nematodes , with more than 70% of genes participating in the process. Trans-splicing even occurs in viruses such as bacteriophage T4, demonstrating an early origin . In archaea, tRNA generation could occur through trans-splicing, for example, in Thermosphaera aggregans and Nanoarchaeum equitans . Split tRNAs were found in some archaea species , and tRNA half homologs were detected in the genomes of archaea, bacteria, and eukaryotes . Recent studies indicate that small guide RNA could be involved in tRNA splicing . Thus, some split tRNAs are proposed to be transcribed from different loci and trans-spliced to generate mature tRNAs. Intriguingly, SL splicing has a much higher frequency, up to approximately 100% compared with other types, including inter/intragenic splicing events, which are observed mainly in dinoflagellates , euglenozoa , nematodes , and rotifers . In addition, in a recent mega-data study, a total of 1,627 trans-splicing events involving 2,199 genes were identified in insects, which accounts for 1.58% of the total genes . This finding, together with many other studies, provides new evidence against the hypothesis that trans-splicing events are merely splicing noise.
Processing Of Eukaryotic Mrna:
Newly synthesized mRNA is called primary transcription or precursor mRNA. It is quite different from the mRNA that takes part in protein synthesis. Large-scale changes take place in precursor mRNA. These changes are called processing of mRNA. Both 5-end 3-end of mRNA are modified. Non-coding regions are removed by splicing. The changes lead to the formation of mature mRNA which takes part in protein synthesis.
5-end Capping:
At 5-end of mRNA a cap of 7-methylguanosine is added immediately after transcription or during transcription. This is most common and is called a cap. This cap is added in reverse orientation of 5 5 instead of normal 5 3 orientation giving a 5-5 triphosphate bridge. A methyl group is added to the 7th position of terminal guanine. This reaction is catalysed by an enzyme guanylyl transferase. The cap provides stability to mRNA as the exonuclease is unable to act on it.
3-Cleavage and Polyadenylation:
At the 3-end of mRNA a tail consisting of sequence of adenine is added immediately after transcription. This tail is called poly tail. It consists of 80-200 adenine nucleotides. This addition is catalysed by an enzyme poly polymerase or PAP. The tail provides stability to mRNA.
Read Also: Exponential Modeling With Percent Growth And Decay Common Core Algebra 2 Homework