Counting Synonymous And Nonsynonymous Differences
Observed nucleotide differences between the two sequences are classified into four categories: synonymous transitions, synonymous transversions, nonsynonymous transitions, and nonsynonymous transversions. When the two compared codons differ at one position, the classification is obvious. When they differ at two or three positions, there will be two or six parsimonious pathways along which one codon could change into the other, and all of them should be considered. Since different pathways may involve different numbers of synonymous and nonsynonymous changes, they should be weighted differently. and made valuable attempts to weight pathways. They also pointed out that equal weighting of pathways, which is used in later methods such as those of and , biases estimates of toward 1 that is, equal weighting tends to overestimate when < 1 and to underestimate when > 1.
ddt,QP
The number of terms used is determined by a preset accuracy level. The weight, that is, the probability for each pathway, is calculated as the product of the probabilities of all changes involved in the pathway. Pathways involving stop codons are given weight 0. If all pathways involve stop codons , ad hoc decisions have to be made.
Which Genes Undergo Positive Selection In Association With A Host Switch
Of obvious interest would be the identification of which genes actually play key roles in host specificity of the wasps. Crucial to any study of this would be the ability to identify those genes that have changed most significantly when wasps have switched hosts during their evolutionary history. Armed with a wasp phylogeny for which we have associated host information, we should be able to use the ratio of nonsynonymous to synonymous substitutions in viral genes in phylogenetically independent contrasts of wasp lineages that have undergone recent host switches versus those that have not.
Previous selection studies have been conducted on PDV genes of known virulence. As an example, Serbielle et al. conducted a study on PDV cystatin gene evolution in a subset of the wasp genus Cotesia. This study showed that particular BV genes were under strong positive selection, with variations being most strongly selected for in amino acids within the active site of the modeled protein structure. Yet, studies such as this have predominantly focused on pre-specified genes by superimposing PDV genomes of interest onto the wasp phylogeny, it should be possible to detect genes of interest and then investigate their function, essentially reversing the process.
Comparison Of Human And Orangutan Mitochondrial Genes
The concatenated sequences of the 12 protein-coding genes on the H-strand of the mitochondrial genome from the human and the orangutan are compared using different methods. The results are shown in . We also included the method of in the comparison, implemented in X. Xias DAMBE program . ML is applied with different assumptions concerning the transition bias and the codon frequency bias. Note that estimates of the dN/dS ratio vary by up to threefold depending on the method/model used. The pattern is especially revealing for ML estimates under different models. For example, accounting for the transition/transversion rate bias increased the proportion of synonymous sites from 25% to 33%, and increased the dN/dS ratio from 0.093 to 0.130. Accounting further for the codon usage bias decreased the proportion of synonymous sites to 28%, with an estimate of = 0.045. The pattern is the same as that in the consistency analysis and the computer simulation discussed above. The results demonstrate that the estimation method is important for estimating dN and dS from real data and that methods accounting for both the transition bias and base/codon frequency bias should be used. This conclusion is consistent with , who found that none of the approximate methods he examined performed well when base/codon frequencies were extreme.
Read Also: What Does Altogether Mean In Math
Where Do Mutations Occur
Mutations can be grouped into two main categories based on where they occur:somatic mutations and germ-line mutations. Somaticmutations take place in non-reproductive cells. Many kinds of somatic mutationshave no obvious effect on an organism, because genetically normal body cellsare able to compensate for the mutated cells. Nonetheless, certain othermutations can greatly impact the life and function of an organism. For example,somatic mutations that affect cell division are the basis for many forms of cancer.
Germ-linemutations
Absolute Dating Based On Synonymous Substitutions
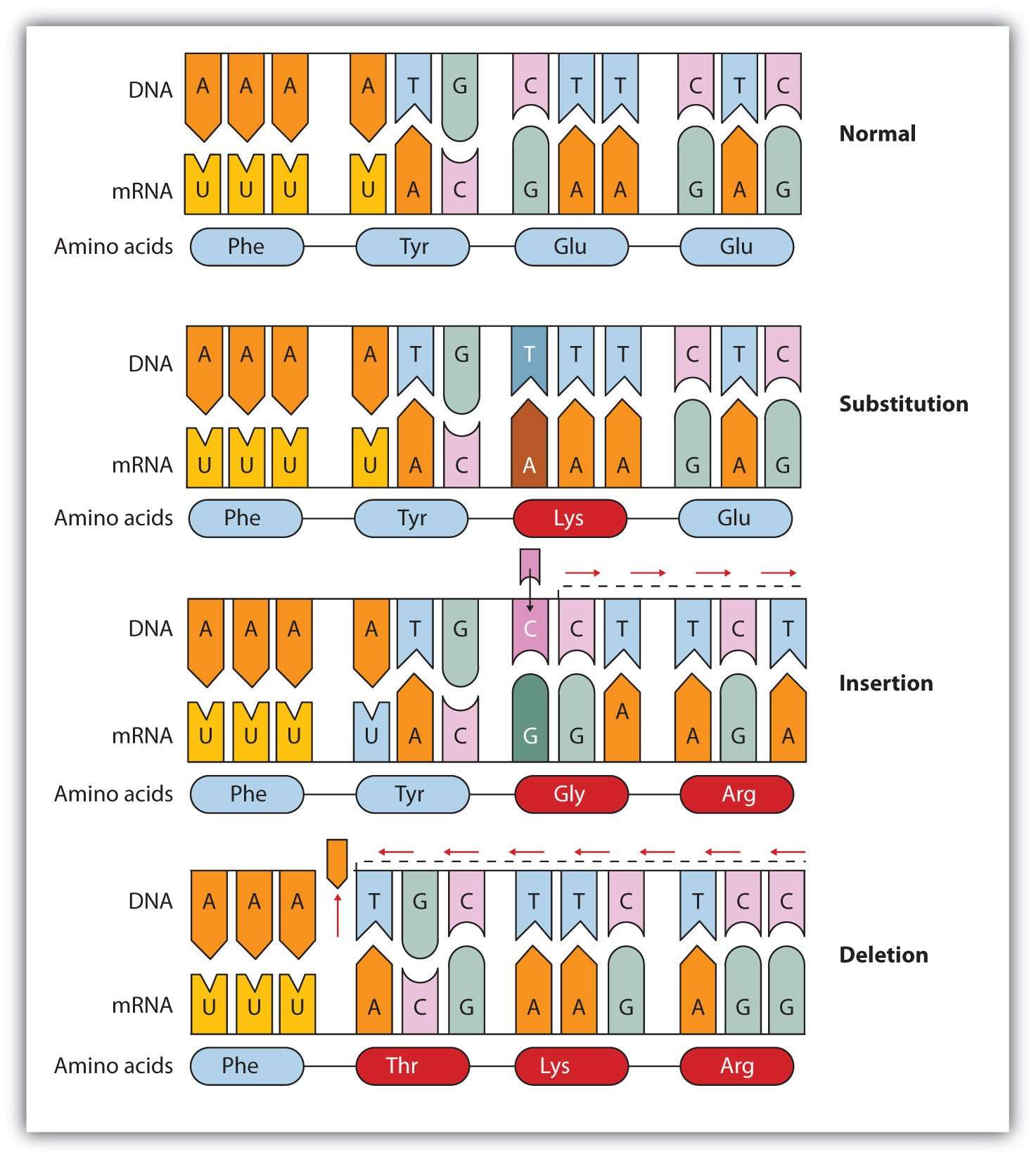
Because most substitutions in third-codon positions do not result in amino acid replacements , the rate of fixation of these substitutions is expected to be relatively constant in different protein-coding genes and to reflect the overall mutation rate . Time of divergence can be calculated from this as T = KS/2, where KS is the fraction of synonymous substitutions per synonymous site and is the mean rate of synonymous substitution . The value for differs for various organisms in Arabidopsis, for instance, the estimate is 6.1 synonymous substitutions per 109 years, whereas for mammals it is considered to be about 2.5 substitutions per 109 years .
FIGURE 6.11. Silent substitutions, indicated in bold, mostly occurring at third codon positions, do not lead to amino acid replacement and are therefore regarded as neutral, and assumed to follow a clocklike behavior.
T. Ohta, in, 2001
Recommended Reading: Glencoe Mcgraw Hill Geometry Concepts And Applications Answers
What Causes A Substitution Mutation
A substitution mutation can be caused by a number of sources directly related to the reading and storage of DNA. For instance, every hour each cell in your body losses around 1,000 nucleotides from the DNA backbone. These nucleotides fall off due to the process of depurination. In the process of replacing them, the proteins that manage the DNA make a mistake approximately 75% of the time, because there are 4 nucleotides to choose from. Other proteins must come along after and check the DNA for errors. If they miss the substitution mutation, it may stay and be replicated.
Another factor which can drive a substitution mutation is deamination, the process by which amino groups degrade off of nucleotides. One of the only ways the protein machinery can differentiate between nucleotides is the amino groups attached to them. As these fall off, the protein machinery can misrecognize the nucleotide, and supply the wrong nucleotide pair. When the DNA replicates, the new nucleotide will become established in a new cell line.
On top of these internal drivers which can cause a substitution mutation, there are also external forces which can cause nucleotide swaps. Carcinogens and mutagens are a special classes of chemicals which drastically impede the protein machinery and cause lots of mutations. Even sunlight can degrade and impede with DNA function, driving a substitution mutation.
Survey Of Mitochondrial Sequence Divergence
Similarity searches were performed with blastn against the nonredundant GenBank database using the atp1, cox1, and matR genes from Arabidopsis thaliana as queries . Limitations were enforced during the searches such that only hits from the Spermatophyta with an e-value less than 1e-10 were included. Hits less than 200 bp in length, multiple hits to the same species, pseudogenes, and cDNA sequences were removed from the blast results, while sequences generated in this study were added. Sequences were aligned using ClustalX and manually adjusted when necessary. Poorly alignable regions were excluded from the data sets, as were regions with gaps in the majority of taxa. These data sets are available .
To determine whether the PAUP* time-saving strategies had an effect on the results of this study, a ML topology was also determined for each gene using RAxML version 2.2.3 . In contrast to the PAUP* analysis, no topological constraints were enforced, the GTR+G model was used , the search started from a maximum parsimony tree, and the ML parameters were estimated during the ML analysis. For each gene, 10 replicates were performed starting from 10 different randomized parsimony trees. The initial rearrangement setting was fixed to 10 for all analyses.
You May Like: Geometry Dash World How To Get Keys
What Is Addition Mutation In Biology
insertionadditioninsertioninsertion
. Keeping this in view, what can insertion mutation cause?
An insertion mutation is a permanent change in a DNA sequence caused by the addition of nucleotides. If the number of nucleotides inserted is a multiple of three, then it is a non-frameshift mutation. Any insertion mutation that causes a nonfunctional protein could result in a disease.
Also Know, what is deletion mutation in biology? In genetics, a deletion is a mutation in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome.
Moreover, what are the types of mutation in biology?
There are three types of DNA Mutations: base substitutions, deletions and insertions. Single base substitutions are called point mutations, recall the point mutation Glu —–> Val which causes sickle-cell disease. Point mutations are the most common type of mutation and there are two types.
What is the mutation caused by the addition of a nucleotide?
Answer Expert Verified. Insertion is a type of mutation which adds nucleotide to an already existing gene sequence. It can lead to genetic disorders due to the change of gene sequence. An example of this genetic disorder is the trisomy 21 or commonly known as Down syndrome.
S For Estimating Synonymous And Nonsynonymous Ratesmaximum
Parameter is the transition/transversion rate ratio, and = dN/dS is the nonsynonymous/synonymous rate ratio. The equilibrium codon frequencies are calculated using the nucleotide frequencies at the three codon positions that is, codon frequencies are proportional to the products of nucleotide frequencies at the three codon positions. This approach was found to produce results similar to those obtained using all codon frequencies as free parameters, although codon frequencies are often quite different from those expected from nucleotide frequencies at codon positions . We note that different assumptions about codon frequencies can be easily incorporated into the ML method and the approximate method of this paper. Equation is similar to the simulation model of and the likelihood model of , although those authors did not consider the transition/transversion rate bias or different base frequencies at the three codon positions.
Qtpijijt.ijnijijit,ddijii.ttdtdt
Don’t Miss: Cpm Algebra 2 Answers
Ranking Genes In Cancer Of Each Tissue And Ranking Cancer Tissues For Each Gene
Supplementary Methods pzNNpSupplementary Table S1Open in new tab
Histograms showing the frequency of occurrence of each of the 12 possible base changes at pos1, pos2 and pos3 of codons in: synonymous missense and nonsense substitutions. In each histogram, base changes are indicated along the x -axis, the number of times that each base change is observed is indicated along the y -axis and the frequencies of base changes at pos1, pos2 and pos3 of codons are shown as separate series.
Correcting For Multiple Hits Using Estimated Numbers Of Sites And Differences
We use the distance formula for the F84 model to correct for multiple substitutions at the same site to calculate dS and dN. For each of the synonymous and nonsynonymous site classes, the proportions of transitional and transversional differences are calculated separately to give T and V for use in equation . Different base frequencies are also used for the two site classes. The correction formula used here, as well as those used in all previous approximate methods, is ad hoc. The formula is derived from a Markov process of nucleotide substitution with four states, where each nucleotide can change into one of three other nucleotides. When the formula is used for synonymous sites only, this basic assumption of the Markov model is violated . However, a proper treatment of the evolutionary process at synonymous and nonsynonymous sites appears to require the use of a codon substitution model, as in the likelihood method, and an analytical derivation of a correction formula based on such a model seems intracTable.
Also Check: Beth From Child Of Rage Now
What Kinds Of Gene Variants Are Possible
The DNA sequence of a gene can be altered in a number of ways. Gene variants can have varying effects on health, depending on where they occur and whether they alter the function of essential proteins. Variant types include the following:
Substitution
This type of variant replaces one DNA building block with another. Substitution variants can be further classified by the effect they have on the production of protein from the altered gene.
- Missense: A missense variant is a typeof substitution in which the nucleotide change results in the replacement of one protein building block with another in the protein made from the gene. The amino acid change may alter the function of the protein.
- Nonsense: A nonsense variant is another type of substitution. Instead of causing a change in one amino acid, however, the altered DNA sequence results in a stop signal that prematurely signals the cell to stop building a protein. This type of variant results in a shortened protein that may function improperly, be nonfunctional, or get broken down.
Insertion
An insertion changes the DNA sequence by adding one or more nucleotides to the gene. As a result, the protein made from the gene may not function properly.
Deletion
Deletion-Insertion
Duplication
A duplication occurs when a stretch of one or more nucleotides in a gene is copied and repeated next to the original DNA sequence. This type of variant may alter the function of the protein made from the gene.
Inversion
Frameshift
Repeat expansion
What Is An Example Of A Substitution Mutation
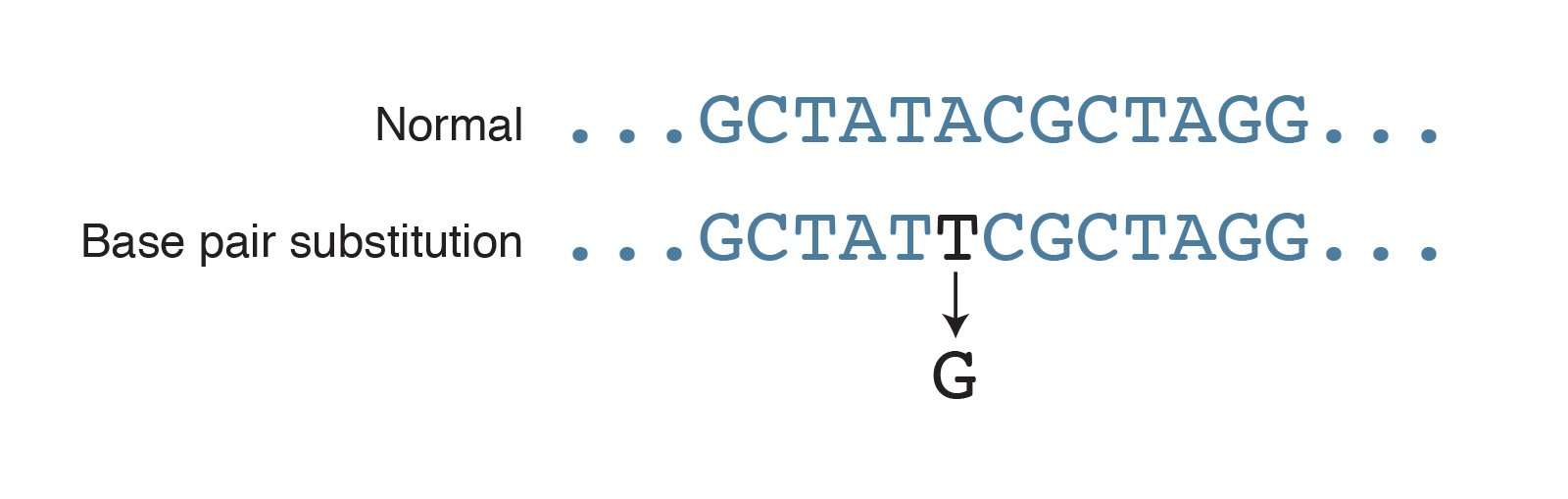
Such a substitution could: change a codon to one that encodes a different amino acid and cause a small change in the protein produced. For example, sickle cell anemia is caused by a substitution in the beta-hemoglobin gene, which alters a single amino acid in the protein produced.
Explanation:
For example, sickle cell anemia is caused by a substitution
answer i believe the correct answer is
Recommended Reading: Why Are Michael Jackson’s Kids White
Patterns Of Molecular Evolution
As compared with the rate of phenotypic evolution, the rate of molecular evolution is remarkably constant, that is, a molecular clock is often observed. This characteristic of molecular evolution is strong evidence for the neutral theory as it is explained by eqn . Another characteristic is the fact that the more constrained the protein is, the lower is its rate of evolution. This is also thought to support the neutral theory, since the proportion of neutral mutations is thought to decrease as the functional constraint becomes stronger. Through comparative studies of DNA sequences, many interesting patterns of evolution and polymorphism have emerged. The difference in average patterns between synonymous and nonsynonymous substitutions of mammalian genes is in accord with selective near-neutrality, that is, the generationtime effect is usually larger for the synonymous substitutions than for the nonsynonymous substitutions.
Real difficulty lies in distinguishing the near-neutrality from the selection theory. The choice is not one or the other, and there may be cases in which both selection and drift are almost equally important. It would be unwise to decide which of the theories is correct in such cases.
David H. Spencer, … John Pfeifer, in, 2015
Counting Synonymous And Nonsynonymous Sites
for counting synonymous and nonsynonymous sites in each codon is correct for mutation models more general than that of , although Inas Table involves minor errors for codons that can change to stop codons in one step. In general, synonymous and nonsynonymous sites can be counted as in the ML method discussed above for any codon-substitution model . We count sites using codons in the two compared sequences, rather than the equilibrium codon frequencies expected from the model . As there should be about 4% loss of sites due to mutations to stop codons, this scaling means that we are slightly underestimating dS and dN, although the ratio is not affected . The numbers of sites are scaled so that S + N = 3Lc, where Lc is the number of codons. Nucleotide frequencies at synonymous and nonsynonymous sites are recorded and used later for multiple-hit corrections.
Don’t Miss: Abiotic Meaning In Science
What Are Insertion Mutations
Insertion mutations are mutations caused due to the addition of one or more nucleotides into a DNA sequence. Therefore, extra nucleotides are added to a DNA sequence in the insertion mutation. Similar to deletion mutations, insertion mutations can be small or large, depending on the number of base pairs added. In small insertions, a single base pair is generally added. Generally, in large insertion mutations, a small fragment of a chromosome is added newly.
Figure 02: Insertion Mutation
Insertion of one base pair can cause a frameshift mutation changing the entire codon sequence. Insertion of three base pairs may be less harmful than the insertion of a single base pair. This is because three base pair insertion does not cause a frameshift mutation. Insertion of a large piece of DNA can be detrimental. If a stop codon is inserted accidentally during an insertion mutation, it leads to a premature end to translation resulting in a non-functional short protein.
Hiv Env Codon Frequencies
shows estimates of when base/codon frequencies from the HIV envelope genes are used. Base frequencies in this gene are less biased than are those in the mitochondrial genes, and the effect of ignoring the base frequency bias is minor. For example, the correct proportion of synonymous sites at = 1 is 21.9%, while NG gives 24.0%, with very slight overestimation. Patterns in are quite similar to those for equal codon frequencies . Exactly at = 1, NG gives the estimates 1.105, 0.371, and 2.554 when the true = 1, 0.3, and 3, respectively. The estimates are biased toward 1, mainly due to the use of equal weighting in counting differences. When = 10, NG underestimates the proportion of synonymous sites . The bias is not as extreme as that in the case of equal codon frequencies, as unequal base/codon frequencies appear to counterbalance the effect of transition bias to some extent .
method overestimates the ratio because it ignores the base frequency bias and thus overestimates the number of synonymous sites. The bias is not as extreme as it is for mitochondrial genes. The new method gives estimates very close to the true values for = 1 and = 0.3. When = 3, the new method slightly underestimates the ratio, as in the case of mitochondrial codon frequencies.
Also Check: Holt Geometry Chapter 7 Test Form C Answers