Effects Of Essentiality In Operon Conservation
Essential genes are usually defined as those whose inactivation prevents growth even in rich medium . The operons of E. coli overrepresent essential genes and those encoding them are highly conserved . Small genomes, which we have shown have more conserved operons, have a larger fraction of essential genes. We have thus tested the hypothesis that essentiality plays a role in the association between operon conservation and genome size. We classed genes as essential or nonessential using E. coli data , and tested whether pairs of essential genes are more often conserved in the same operon than pairs of nonessential genes . This was true and statistically significant in all clades , with stronger effects in -Proteobacteria, then -Proteobacteria and finally Firmicutes. We then tested how the presence of essential genes affected operon conservation in function of genome size. Both pairs of essential and nonessential genes tend to be less conserved in the operons of larger genomes . Yet, the correlation is weak and often nonsignificant. These results not only show that operons with essential genes are more conserved, but also show that both operons with essential and nonessential genes are less conserved in large genomes.
What Happens When Lac Operon Is On
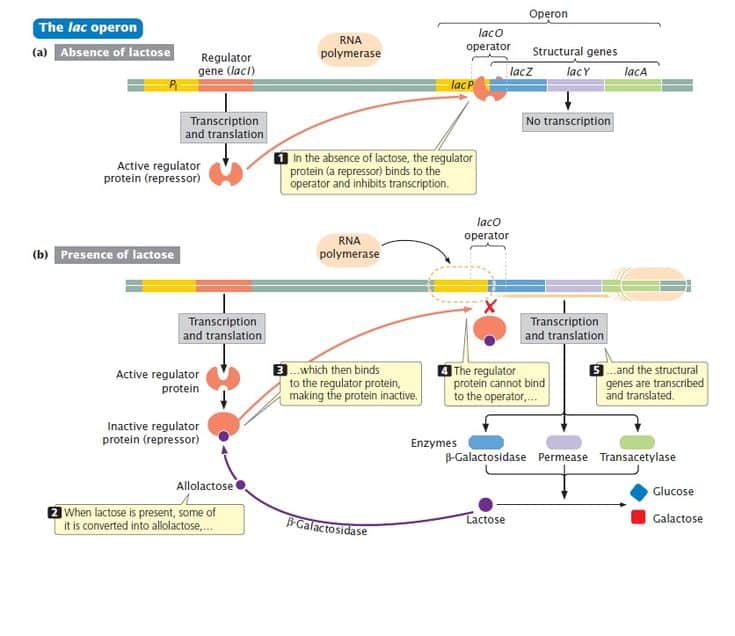
The lac operon of E. Two regulators turn the operon on and off in response to lactose and glucose levels: the lac repressor and catabolite activator protein . The lac repressor acts as a lactose sensor. It normally blocks transcription of the operon, but stops acting as a repressor when lactose is present.
Associations Between General Features Of Operon Organization And Genome Features
We used two independent databases of operon predictions, ProOpDB and DOOR, to class pairs of adjacent genes as intraoperonic or interoperonic . These analyses were done for 124 genomes of Firmicutes, 102 of – and 63 of -Proteobacteria . The analysis of both operon databases gave concordant results and we only present in the main text the results concerning ProOpDB, which reports the highest accuracy . Species in our data set are not independent statistical instances because they are related by a common evolutionary history. To correct for this effect, we retested all correlation analyses using PICs . For simplicity, we indicate the results of these analyses in the main text only when they qualitatively contradict the significance of the Spearman nonparametric association test.
Association between genome size and the fraction of genes in operons, the length of operons and the density of coding sequences. Association between the fraction of genes in operons and the length of operons as a function of genome size . Association between the fraction of genes in operons and the density of coding sequences . Results for -Proteobacteria , -Proteobacteria , and Firmicutes . Operon length was calculated as the average number of genes per operon for the whole genome operon predictions. Association between the variables was computed using the nonparametric Spearman rho.
Also Check: Is An Invasive Technology Used To Treat Psychological Disorders
Role Of Cap In Arabinose Operon Regulation:
Catabolite activator protein is a small homodimeric DNA binding protein. It helps to recruit the RNA polymerase to the promoter site. If glucose is present in the cell the cAMP inside the cell decreases due to inactive adenylate cyclase. Enzyme adenylate cyclase synthesizes the cAMP . This enzyme is negatively regulated by glucose transport.
So high glucose levels will block the function of adenylate cyclase and cAMP levels drop due to inactive adenylate cyclase and binding of cAMP to CAP site is not possible. In contrast, if the glucose levels are low, high levels of cAMP bind to the CAP site forming the cAMP-CAP complex and providing additional stimulation to the RNA polymerase and enhancing the efficiency of transcription.
Glucose Arabinose cAMP CAP inactive Negative regulation
Glucose Arabinose cAMP CAP active Positive regulation
This Arabinose operon is a unique example of an operon where the regulatory protein acts as a repressor when arabinose is absent and as an activator when arabinose is present.
Hope you liked this articleabout Arabinose Operon. If you think there is something in this article or needs any kind of improvement you can let us know in the comment box below. After reading the gene expression in phages you will also like other articles on molecular biology.
The Trp Operon: A Repressor Operon
Bacteria such as E. coli need amino acids to survive. Tryptophan is one such amino acid that E. coli can ingest from the environment. E. coli can also synthesize tryptophan using enzymes that are encoded by five genes. These five genes are next to each other in what is called the tryptophan operon . If tryptophan is present in the environment, then E. coli does not need to synthesize it and the switch controlling the activation of the genes in the trp operon is switched off. However, when tryptophan availability is low, the switch controlling the operon is turned on, transcription is initiated, the genes are expressed, and tryptophan is synthesized.
E. colitrp
A DNA sequence that codes for proteins is referred to as the coding region. The five coding regions for the tryptophan biosynthesis enzymes are arranged sequentially on the chromosome in the operon. Just before the coding region is the transcriptional start site. This is the region of DNA to which RNA polymerase binds to initiate transcription. The promoter sequence is upstream of the transcriptional start site each operon has a sequence within or near the promoter to which proteins can bind and regulate transcription.
Don’t Miss: What Does L Stand For In Physics
Analysis Of Expression Levels
To estimate E. coli K-12 MG1655 mRNA levels per cell, we used four public transcriptomic data sets . Data obtained from different experiments were normalized. For this, we selected the genes that were assayed in all experiments . We computed the average values for these genes in each experiment. If in experiment A the average is XA and in experiment B it is 1.3*XA, then 1.3 is the multiplicative factor that has to be applied to the values in A so that these genes can be compared with the ones of B. The ratio of the averages between experiments gives then the multiplicative factors that allow us to normalize the experiments so that we can compare them directly. In the earlier example, a gene z assayed in A and B has a normalized value of /2. To estimate the mRNA levels of each OGP, we averaged the normalized values of both genes. Then, we used the median of this distribution to divide the set of genes in two equal size subsets of highly expressed and LE genes.
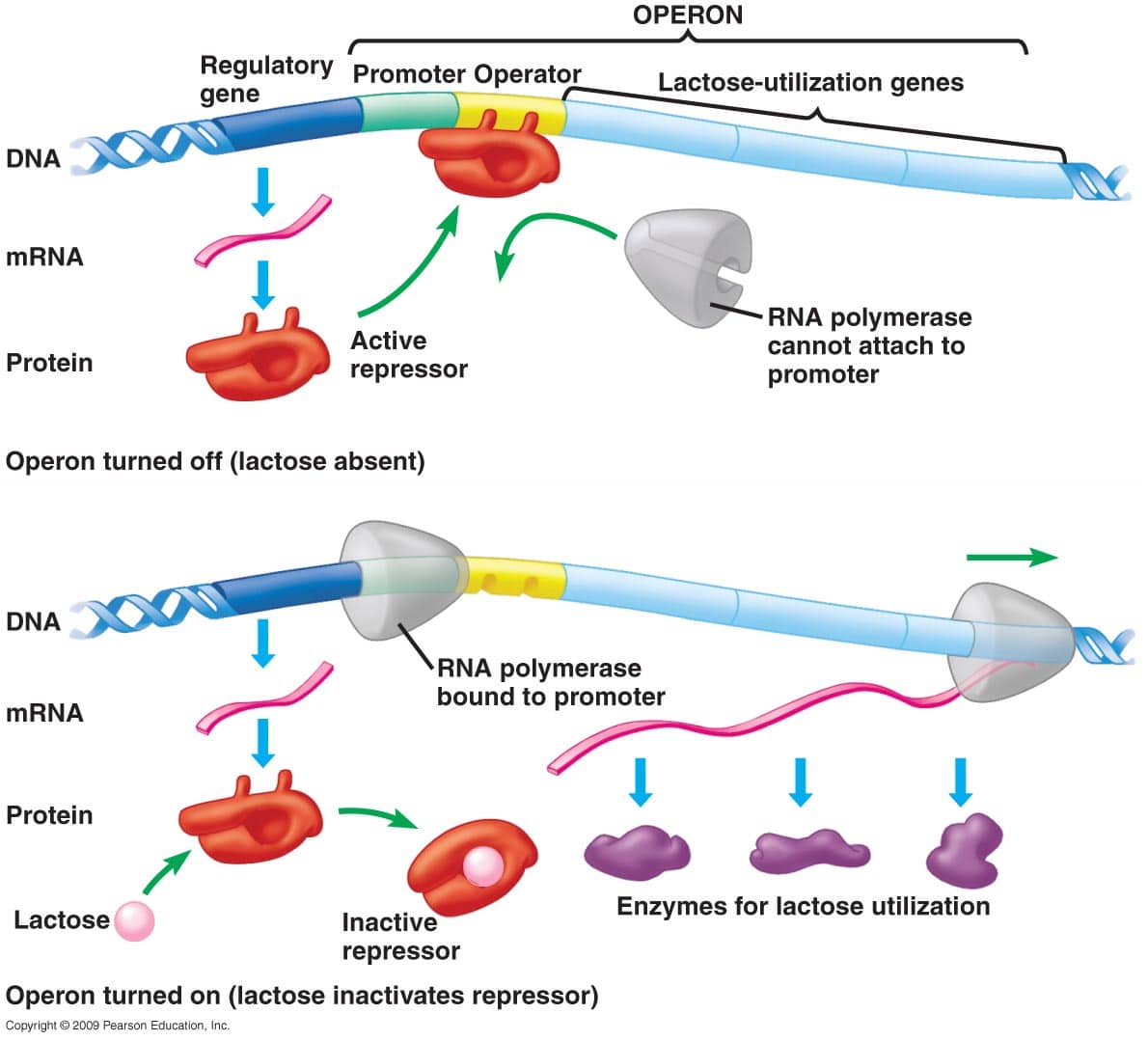
Operons: Description And Definition
Many of E. colis genes are organized into systems called operons. Operons consist of the following components.
The DNA at 4 consists of structural genes. These are the genes that code for enzymes or other gene products. These structural genes are under the control of a regulatory gene . The regulatory gene produces a regulatory protein that determines whether or not the structural genes will be transcribed into mRNA, which in turn is translated into protein. Because the regulatory protein often has the effect of preventing transcription, the regulatory protein is often referred to as a repressor.
Transcription is carried out by RNA polymerase, which can only transcribe a gene by binding with a region of DNA called the promoter, shown above at 2. Just downstream of the promoter is a region called the operator . The operator is a binding site for the regulatory protein/repressor that the regulatory gene codes for. Pulling this together, we can define an operon as follows :
a cluster of genes under the control of a single promoter. The genes are transcribed and regulated in such a way that these genes are either expressed together or not at all.
This will become clearer by looking at an operon in action. Well start with the lac operon, the first operon whose function was understood. This understanding came about through the work of François Jacob, André Michel Lwoff, and Jacques Monod, who received the Nobel Prize for their work in 1965.
Don’t Miss: What Is Clinical Observation In Psychology
What Is An Operon
What is an operon in a eukaryotic cell, and how does it regulate the expression of genes? I’ve already read Wikipedia, but it is not enough clear to me. Unfortunately my knowledge in genetics are very poor!
- HDE 226868Dec 29, 2014 at 13:55
- $\begingroup$There are some naturally occurring polycistronic mRNAs in flies and worms. You can also make artificial polycistronic DNA using IRES sites or self-cleaving 2A peptide sequences . Which one are you asking?$\endgroup$Dec 29, 2014 at 16:31
- 1$\begingroup$Is an operon just several genes with the same regulation, so they are transcribed at the same time? In prokaryotes the genes in an operon are often placed next to each other so they can be transcribed as one mRNA and take advantage of their ability to translate polycistronic mRNA, but in eukaryotes where polycistronic mRNAs are less common, they might not need to be near each other in the genome.$\endgroup$Dec 29, 2014 at 17:19
- $\begingroup$@Superbest: I’m asking about naturally occurring polycistronic mRNAs$\endgroup$
Operons were once thought to appear only in prokaryotes , but there are now known to be a number examples in eukaryotic organisms. These are mostly seen in nematodes and insects.
In prokaryotes the expression of the operon leads to polycistronic mRNA, which is then translated into proteins.
Operons in eukaryotes are also gene clusters , which are regulated by one regulatory unit . See figure 1 from reference 1:
References:
Control Of Gene Expression In Prokaryotes
In prokaryotes, the Lac-operon system is controlled in two ways:
- Positive control
It is also called a Positive inducible system and includes the following steps:
This concept is known as the switch on of Lac-operon .
Negative Control of Lac-Operon
It is also called Negative control of the repressor system. It includes the following steps:
This concept is known as the switch off of Lac-operon .
Inducer
Recommended Reading: What Is Conscientiousness In Psychology
Analysis Of Transcription Factors
Information for the predicted transcription factors for each genome was downloaded from the DBD transcription factor database . We analyzed all organisms in our study for which there were predictions available of transcription factors: 49 -proteobacteria, 35 -proteobacteria, and 59 Firmicutes. We included in the analysis the nonredundant transcription factor protein families using UCLUST with a threshold of 80% .
Functions Of Gene Clusters In Animal Defense And Development
Global gene expression analysis has revealed extensive clustering of non-homologous genes that are co-ordinately expressed in eukaryotes, including in animals . These groups of genes may be expressed during development, or in certain tissues and diseased states, and have been reported in studies of Drosophila, nematode, mouse, and humans. Such co-expression domains may therefore be an important source for the discovery of new functional gene clusters in animals and other eukaryotes. However, more research is needed before we can fully understand the functional significance of co-expression domains . Of the known functional gene clusters in animals, the best characterized is the major histocompatability complex , which encodes proteins involved in innate and adaptive immunity. Other classes of mammalian gene clusters include the Hox and -globin loci, which are required for development and for the synthesis of haemoglobin, respectively. The latter two examples consist of genes that share sequence similarity and so are distinct from classical operons and from the functional gene clusters discussed above. However, investigation of these loci has revealed important insights into the mechanisms of regulation of arrayed gene clusters in eukaryotes and so these gene clusters will also be considered here.
Recommended Reading: What Is Movement In Geography
Interactions Between Operator And Repressor
Sequence of operator
The operator overlaps the start the site of transcription and the promoter. It has a dyad symmetry centered at +11. Such a dyad symmetry is commonly found within binding sites for symmetrical proteins . The sequence of DNA that consititutes the operator was defined by the position of oC mutations, as well as the nucleotides protected from reaction with, e.g. DMS, upon binding of the repressor.
a. Purification
Increase the amount of repressor in the starting material by over-expression.
A wild-type cell has only about 10 molecules of the repressor tetramer. Isolation and purification of the protein was greatly aided by use of mutant strain with up-promoter mutations for lacI, so that many more copies of the protein were present in each cell. This general strategy of over-producing the protein is widely used in purification schemes. Now the gene for the protein is cloned in an expression vector, so that the host makes a large amount of the protein – often a substantial fraction of the total bacterial protein.
Assays for repressor
Binding of radiolabeled IPTG to repressor
Binding of radiolabeled operator DNA sequence to repressor. This can be monitored by the ability of the protein-DNA complex to bind to nitrocellulose . Electrophoretic mobility shift assays would be used now in many cases.
b. The isolated, functional repressor is a tetramer each of the four monomers is the product of the lacI gene .
3. Contact points between repressor and operator
Catabolic Versus Biosynthetic Operons
Catabolic pathways catalyze the breakdown of nutrients to generate energy, or more precisely ATP, the energy currency of the cell. In the absence of the substrate,there is no reason for the catabolic enzymes to be present, and the operon encoding them is repressed. In the presence of the substrate, when the enzymes are needed, the operon is induced or de-repressed.
| Operon encodes |
|---|
lacZ encodes b-galactosidase, which cleaves the disccharide lactose into galactose and glucose.
lacYencodes the lactose permease, a membrane protein that faciltitates uptake of lactose.
lacAencodes b-galactoside transacetylase the function of this enzymes in catabolism of lactose is not understood
Recommended Reading: Best Mice For Geometry Dash
Operon Conservation And Genome Size
We computed for each genome the OCI . OCI is the fraction of intraoperonic pairs of adjacent genes in a genome having orthologs in one same operon in E. coli. OCI values are higher in -proteobacteria as expected because this is the clade closest to E. coli. The other clades have lower medians and their respective ranksFirmicutes and -proteobacteria are opposite to the expected given phylogenetic relatedness. Next, we examined the association between genome size and OCI. The three clades showed negative, statistically significant, correlations between operon conservation and genome size . Hence, larger genomes have fewer, shorter and less conserved operons than smaller genomes. To shed some light in the gene traits shaping operon conservation, we introduced in our analysis a series of biologically relevant variables such as gene essentiality, protein expression levels, and balanced protein concentration.
Association of the OCI with genome size. -Proteobacteria -Proteobacteria and Firmicutes. Association between the variables was computed using the nonparametric Spearman rho.
Positive Regulation: Glucose & Arabinose
When the arabinose is present for bacteria to consume naturally we would expect that the arabinose operon is activated which means the araBAD expression is upregulated. This regulation is driven by the change in the structure of the ara C protein. Typically, the ara C monomer has an N-terminal domain, C-terminal domain, and linker. When the arabinose is present in bacteria it can directly bind to the arabinose binding pocket of the N-terminal domain of ara C.
This binding causes a conformational change in the ara C monomer where the N-terminal domain arm flips out and acts like a lid for the arabinose binding pocket. This lid stabilizes the arabinose in the pocket.
In the default or non-active state, the DNA looping represses the araBAD expression but when arabinose is present it binds to the ara C dimer and causes the conformational changes in ara C. This changes the topology of the DNA by destabilizing the DNA loop. After the loop is destabilized the ara C binds to the I1 and I2.
In the off-state, the ara C dimer can only have long-distance contacts through DNA looping but when the arabinose is bound it changes the ara C dimer configuration so that it only forms short-range contacts. In the upstream region of the inducer there is an operator site O1. So the operator site can also have the ara C dimer bound to it. The arabinose bound to ara C at ara I sites interacts with RNA polymerase and with the help of CAP it promotes strong activation of araBAD expression.
Also Check: What Are Fundamental Quantities In Physics
Catabolite Activator Protein : An Activator Regulator
Just as the trp operon is negatively regulated by tryptophan molecules, there are proteins that bind to the operator sequences that act as a positive regulator to turn genes on and activate them. For example, when glucose is scarce, E. coli bacteria can turn to other sugar sources for fuel. To do this, new genes to process these alternate genes must be transcribed. When glucose levels drop, cyclic AMP begins to accumulate in the cell. The cAMP molecule is a signaling molecule that is involved in glucose and energy metabolism in E. coli. When glucose levels decline in the cell, accumulating cAMP binds to the positive regulator catabolite activator protein , a protein that binds to the promoters of operons that control the processing of alternative sugars. When cAMP binds to CAP, the complex binds to the promoter region of the genes that are needed to use the alternate sugar sources . In these operons, a CAP binding site is located upstream of the RNA polymerase binding site in the promoter. This increases the binding ability of RNA polymerase to the promoter region and the transcription of the genes.
E. coli